In this Part 2 of a series I again delve into some technical detail, but this time in an effort to catch a little bit of the form and movement, the dynamism, of genetic and epigenetic processes. I well realize that the material in this series will present a challenge to many readers. The presentation is both denser and lengthier than normal for NetFuture. I've been motivated to produce these articles by the awareness that epochal changes in biology are underway, and that the popular media have so far grossly failed to give the public any realistic feel for what is happening. A reasonably detailed and generally accessible summary of the ongoing, highly technical research needs to be available somewhere.
The readership for such material may be rather more limited than for the general run of NetFutures, but I consider this readership extremely important. And you can help me out in one regard: if you find these articles of use, please drop me a line to let me know. I will also welcome any suggestions for improvement.
I am currently expecting that there will be a total of four articles tracing some of the current research relating to epigenetics and gene regulation. These will be followed by attempts to draw out the significance of the work for biology, for science in general, and for the controversial notion of "holism". I do believe we are headed toward a revision of the scientific outlook that may be the equal of any of the great "paradigm shifts" of the past — this despite the fact that penetrating discussion of the implications of the current work has scarcely even begun. It's almost as if no one — or, at least, no one who is attempting to publish in the primary, peer-reviewed journals — has quite dared as yet to step outside the old rhetoric of "genetic code", "mechanism", "building blocks", and all the rest, and actually face what is going on.
On a different note: if you are interested in evolution, you might want to check out two articles in the latest issue of The Nature Institute's hardcopy newsletter, In Context. These articles have been posted to the web.
SLT
(You will find the latest versions of the currently available parts of this series at the website, "From Mechanism to a Science of Qualities".)
By clicking on the shaded rectangles at the end of many scientific terms, you can immediately read a definition of the terms in a separate window. This requires JavaScript to be enabled in your browser.
The decades following the 1953 discovery of the double helix![]() were a time
when everything important seemed to hinge on the fixed and definitive
"genetic code"
were a time
when everything important seemed to hinge on the fixed and definitive
"genetic code"![]() . The
researcher's task was to work out explicitly a prescriptive logic already
contained in the code. It was not a time when molecular biologists were
likely to preface their discussions of gene expression
. The
researcher's task was to work out explicitly a prescriptive logic already
contained in the code. It was not a time when molecular biologists were
likely to preface their discussions of gene expression![]() with
statements like this:
with
statements like this:
DNA is a living molecule, writhing, twisting and bending in response to the physical forces applied to it by genetic processes (Kouzine et al. 2007).Nor was it a time when gene packaging materials and a diverse bestiary of regulatory factors
Or, again, as recently as 1996, when the yeast genome was fully decoded and all its secrets supposedly laid bare, who would have expected to read this a decade further on:
Even for a [yeast] genome that has been studied intensively since it was sequenced10 years ago, a glimpse into the complexity of its transcriptional
architecture makes this genome appear like novel territory (Lior 2006 et al.).
How could the relatively simple yeast genome remain novel ten years after the complete unveiling of its structural sequence? And as for the choreography of the cellular "dance", isn't this a mere metaphor? Surely the authors quoted above did not really mean to say that a dance controls the genome! After all, the decisive logic of the genomic code is supposed to be what ultimately controls everything else.
Or so it has been thought. But the pace of discovery today is almost blinding, and it seems to be a time for the loosening of old thought patterns. The appeals to genomic plasticity, balance, tension, and context, the language of dynamism and artistic movement — these are not mere signs of a collective weakness for poetic invention, but rather are evoked by the phenomena under consideration. There is, in fact, nothing to prevent the contemporary molecular biologist from thinking along the following lines:
The organism must be rooted in something more than an abstract, fixed, and unchanging logic. It is a dynamic material presence, and requires a materially effective genesis. If DNAand all the other contents of the living cell provide the physical foundation for the organism as a whole — for the seeing eye, the beating heart, the graceful and compelling movement of the ballet dancer — then why shouldn't their own, molecular performance be at least as artful and complex as that which they support on a larger scale? Why shouldn't their gestures be as meaningful and expressive as those of the organs whose development they so effectively underwrite? Is there any reason to think that the life animating the cell and its DNA should be any more reducible to a static logical sequence — should be any less subtle or capable or dynamic — than eye, heart, and ballerina? Why should we project tiny, neatly programmed mechanisms into the cell when the organisms we see don ’t at all look or behave like such mechanisms?
Of course, these thoughts, especially if expressed in this way, would still today appear rather eccentric. There are, as we will see, logic-centered and mechanistic habits of thought that fiercely oppose giving embodied (and therefore observably aesthetic) form and movement their due.
But nevertheless, the landscape on which we all must move with our ideas,
outworn or forward-looking as they may be, is inexorably changing. It's a
landscape upon which, less than a decade after the height of the fever
induced by the Human Genome Project, the author of a review in a major
biological journal could write that genome![]() mappings and
genomic comparisons of species "shed little light onto the Holy Grail of
genome biology, namely the question of how genomes actually work" in
living organisms (Misteli 2007).
mappings and
genomic comparisons of species "shed little light onto the Holy Grail of
genome biology, namely the question of how genomes actually work" in
living organisms (Misteli 2007).
And so today whoever would take a researcher to task for eccentric thinking might have a harder time of it than usual. In surveying contemporary molecular biology, the least anyone can say is, "Things are getting very interesting!"
If you arranged the DNA![]() in a human
cell linearly, it would extend for about two meters. How do you pack all
that DNA into a cell nucleus about ten millionths of a meter in diameter?
According to the usual comparison it's as if you had to pack 24 miles (40
km) of extremely thin thread into a tennis ball. Moreover, this thread is
divided into 46 pieces (individual chromosomes
in a human
cell linearly, it would extend for about two meters. How do you pack all
that DNA into a cell nucleus about ten millionths of a meter in diameter?
According to the usual comparison it's as if you had to pack 24 miles (40
km) of extremely thin thread into a tennis ball. Moreover, this thread is
divided into 46 pieces (individual chromosomes![]() ) averaging,
in our tennis-ball analogy, over half a mile long. Can it be at all
possible not only to pack these into the ball, but also to keep them from
becoming hopelessly entangled?
) averaging,
in our tennis-ball analogy, over half a mile long. Can it be at all
possible not only to pack these into the ball, but also to keep them from
becoming hopelessly entangled?
Let's begin visualizing the situation in a little more detail.
Imagine you have a rope consisting of two strands spiraling around each
other in the manner of a double helix![]() . Imagine
further that there are short, fairly rigid rods connecting the two strands
at regular intervals along their entire length. There are many of these
rods — roughly ten along the brief length required for one strand to
spiral completely around the other a single time. Each rod represents two
linked nucleotide bases
. Imagine
further that there are short, fairly rigid rods connecting the two strands
at regular intervals along their entire length. There are many of these
rods — roughly ten along the brief length required for one strand to
spiral completely around the other a single time. Each rod represents two
linked nucleotide bases![]() , or base
pairs
, or base
pairs![]() (complementary "letters" of the genetic code
(complementary "letters" of the genetic code![]() ), with one
base bound tightly to one strand of the rope and the other bound to the
complementary strand.
), with one
base bound tightly to one strand of the rope and the other bound to the
complementary strand.
It would be good if the rope, in its double helical form, has been soaked
in a starch solution or some other stiffening agent. This is because DNA,
as a result of its chemical constitution, possesses a degree of natural
rigidity; left to itself, the overall structure resists bending, and one
strand "wants" to wrap around the other a fixed number of times for any
given length of the helical axis![]() . It is
possible to increase this number somewhat (that is, to wind the strands
more tightly around each other) or else to decrease it (unwind the
strands). But in both cases this is to work against the stiffness of the
natural structure, and therefore to create tension that must be
accommodated in one way or another.
. It is
possible to increase this number somewhat (that is, to wind the strands
more tightly around each other) or else to decrease it (unwind the
strands). But in both cases this is to work against the stiffness of the
natural structure, and therefore to create tension that must be
accommodated in one way or another.
You have doubtless seen this accommodation many times. Hold the ends of a
two-stranded rope in your hands and begin to twist one of the ends so as
to tighten the spiraling strands. Before long you will find the rope
coiling into something like a figure eight, and then into ever more
complex forms as you continue twisting, until finally you end up with a
"nest of worms". And much the same happens if you twist in the opposite
direction, as if you were trying to unwind or loosen the spiraling
strands. The coil resulting from a tightening twist is called a positive
supercoil![]() , while a
loosening twist produces a negative supercoil1.
, while a
loosening twist produces a negative supercoil1.
The forces involved in these deformations can be very large — as you
will have discovered if you have ever tried to coerce a stiff rope through
multiple stages of coiling by twisting its ends, or even if you have found
yourself wrestling with a recalcitrant garden hose while trying to coil it
neatly. Matter can be very resistant! Analogous forces come into play
with chromosomes![]() as well.
as well.
DNA![]() is often in
a negatively supercoiled state to one degree or another — and, as we
will soon see, this is owing to additional reasons beyond the fact that,
in order to fit into its own space in the nucleus, it is coiled, bent,
wound and otherwise structured to an almost unfathomable degree. The
question is how any sort of order is maintained: how is the nest of worms
packaged and managed in every cell of the human body in a manner allowing
the thousands of distinct, individual genes, and perhaps hundreds of
thousands of regulatory sequences
is often in
a negatively supercoiled state to one degree or another — and, as we
will soon see, this is owing to additional reasons beyond the fact that,
in order to fit into its own space in the nucleus, it is coiled, bent,
wound and otherwise structured to an almost unfathomable degree. The
question is how any sort of order is maintained: how is the nest of worms
packaged and managed in every cell of the human body in a manner allowing
the thousands of distinct, individual genes, and perhaps hundreds of
thousands of regulatory sequences![]() , to come to
harmonious expression within the complex life of the larger cell?
, to come to
harmonious expression within the complex life of the larger cell?
The first step in DNA packaging involves tiny "spools" made of histone![]() proteins
— some thirty million of them in the human genome
proteins
— some thirty million of them in the human genome![]() , so that
there can be several hundred thousand or more in a single chromosome
, so that
there can be several hundred thousand or more in a single chromosome![]() . The DNA
double helix
. The DNA
double helix![]() commonly
wraps about two times around this spool, continues on for a short
distance, then wraps around a second spool, and so on. The DNA-enwrapped
spool is called a nucleosome
commonly
wraps about two times around this spool, continues on for a short
distance, then wraps around a second spool, and so on. The DNA-enwrapped
spool is called a nucleosome![]() .
.
This first level of DNA packaging is often described as "beads on a
string". (See third image from left in the figure below.) The DNA and
histone spools, together with numerous other attached proteins and smaller
chemical groups, give an overall, ever-changing form and structure to what
is called chromatin![]() — the
actual material of the chromosome. But with this spooling of the double
helix we have seen only the beginning of the compaction that must occur in
order to fit the chromosome into the cell nucleus. Unfortunately, the
higher-order structures of the compacted chromosome are still little
understood. The spools with their DNA somehow get packed into dense,
three-dimensional arrangements, and this entire arrangement coils further
upon itself beyond anyone's current ability to unravel the details. Such
difficulty, however, rarely hinders the adventurous from offering visual
models. So, for what it is worth, here is a conventional picture showing
several stages in the condensation of a chromosome (it is best viewed at
full screen width):
— the
actual material of the chromosome. But with this spooling of the double
helix we have seen only the beginning of the compaction that must occur in
order to fit the chromosome into the cell nucleus. Unfortunately, the
higher-order structures of the compacted chromosome are still little
understood. The spools with their DNA somehow get packed into dense,
three-dimensional arrangements, and this entire arrangement coils further
upon itself beyond anyone's current ability to unravel the details. Such
difficulty, however, rarely hinders the adventurous from offering visual
models. So, for what it is worth, here is a conventional picture showing
several stages in the condensation of a chromosome (it is best viewed at
full screen width):
![[Images of (1) the double helix; (2) nucleosome; (3)
beads on a string; (4) 30 nanometer fiber; (5) active chromosome; and (6)
the chromosome during cell division.]](Images/chromosome1.jpg)
For credits and permissions, see
http://upload.wikimedia.org/wikipedia/commons/4/4b/Chromatin_Structures.png.
During the cell's normal functioning the chromosome is not as fully
condensed as it is during cell division (the two images at far right).
Nor is it all in one state. Some parts of it — especially the parts
containing many active genes — are in something rather more like the
"beads-on-a-string" form, while other parts may be in the conformation of
the 30-nanometer fiber, and vast regions are in a much more wound-up form.
(Thirty nanometers is 30 billionths of a meter, or about 3 thousandths of
the diameter of a typical cell nucleus.) In general — but with
exceptions — the more compact the chromatin![]() , the less
available are the genes for transcription
, the less
available are the genes for transcription![]() .
.
Another image follows below, this one showing four proposed models —
each viewed from two different angles — for the structure of
chromatin in the 30-nanometer fiber. The models do not show the actual
spools or other proteins, but only the DNA![]() . (The DNA is given as a simple "wire",
without any representation of the two helical strands.) However, you can
see how one spool could be positioned inside each double spiral of DNA.
Then, given the scale of the image, you would need to picture this
arrangement extending linearly for enormous distances, even if only a
small part of a chromosome were represented.
. (The DNA is given as a simple "wire",
without any representation of the two helical strands.) However, you can
see how one spool could be positioned inside each double spiral of DNA.
Then, given the scale of the image, you would need to picture this
arrangement extending linearly for enormous distances, even if only a
small part of a chromosome were represented.
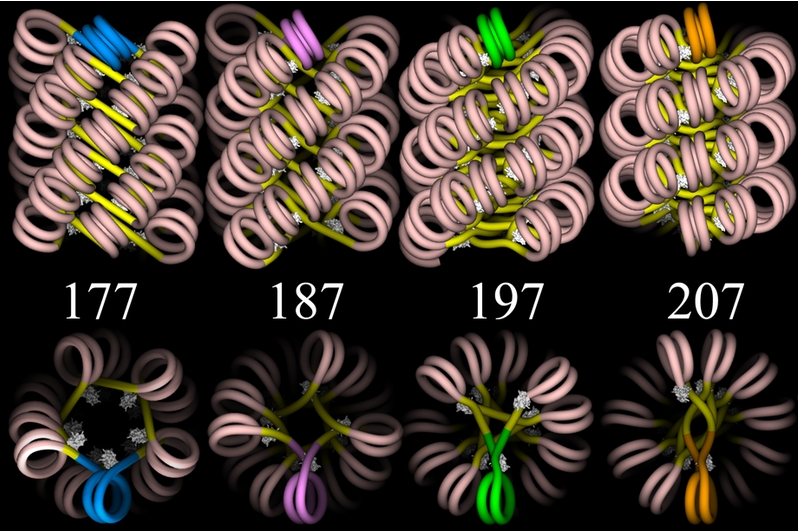 Graphics by Julien Mozziconacci (http://en.wikipedia.org/wiki/File:ChromatinFibers.png) |
Linker DNA![]() — the
short, connecting lengths of DNA between spools — is shown in bright
yellow, and the wrapped DNA is flesh-colored. The different models are
based on different assumptions about the total number of base pairs
— the
short, connecting lengths of DNA between spools — is shown in bright
yellow, and the wrapped DNA is flesh-colored. The different models are
based on different assumptions about the total number of base pairs![]() from the
start of one spool to the start of the next one — that is, the
length of wrapped DNA plus linker DNA. These lengths are the numbers
shown in the figure. You can bring the upper and lower images into proper
relation if you imagine each of the upper images rotated ninety degrees
around a horizontal axis so as to bring the brightly colored (blue, pink,
green, or gold) double spiral of the upper image into the position shown
in the lower image. Finally, the white "lumps" in the figure represent
linker histones
from the
start of one spool to the start of the next one — that is, the
length of wrapped DNA plus linker DNA. These lengths are the numbers
shown in the figure. You can bring the upper and lower images into proper
relation if you imagine each of the upper images rotated ninety degrees
around a horizontal axis so as to bring the brightly colored (blue, pink,
green, or gold) double spiral of the upper image into the position shown
in the lower image. Finally, the white "lumps" in the figure represent
linker histones![]() , which hold
the DNA to the spool and help to stabilize the entire array.
, which hold
the DNA to the spool and help to stabilize the entire array.
Perhaps none of this helps us greatly to understand how the
extraordinarily long chromosome![]() ,
tremendously compacted to varying degrees along its length, can maintain
itself coherently within the functioning cell. But here's one relevant
consideration: there are enzymes called topoisomerases
,
tremendously compacted to varying degrees along its length, can maintain
itself coherently within the functioning cell. But here's one relevant
consideration: there are enzymes called topoisomerases![]() , whose task
is to help manage the forces and stresses within a chromosome.
Demonstrating a spatial insight and dexterity that might amaze those of us
who have struggled to sort out tangled masses of thread, these enzymes
manage to make just the right local cuts to the strands in order to
relieve strain, allow necessary movement of individual genes or regions of
the chromosome, and prevent a hopeless mass of knots.
, whose task
is to help manage the forces and stresses within a chromosome.
Demonstrating a spatial insight and dexterity that might amaze those of us
who have struggled to sort out tangled masses of thread, these enzymes
manage to make just the right local cuts to the strands in order to
relieve strain, allow necessary movement of individual genes or regions of
the chromosome, and prevent a hopeless mass of knots.
Some topoisomerases cut just one of the strands of the double helix![]() , allow it to
wind or unwind around the other strand, and then reconnect the severed
ends. Other topoisomerases cut both strands, pass a loop of the
chromosome through the gap thus created, and then seal the gap again.
(Imagine trying this with miles of string crammed into a tennis ball
— without tying the string into knots!) I don't think anyone would
claim to have the faintest idea how this is actually managed in a
meaningful, overall, contextual sense, but great and fruitful efforts are
being made to analyze isolated local forces and "mechanisms".
, allow it to
wind or unwind around the other strand, and then reconnect the severed
ends. Other topoisomerases cut both strands, pass a loop of the
chromosome through the gap thus created, and then seal the gap again.
(Imagine trying this with miles of string crammed into a tennis ball
— without tying the string into knots!) I don't think anyone would
claim to have the faintest idea how this is actually managed in a
meaningful, overall, contextual sense, but great and fruitful efforts are
being made to analyze isolated local forces and "mechanisms".
Before we try to bring the picture a little more alive, there's one small exercise that may help us. Many window shades have a looped cord for adjusting the light. If you slip your finger through the loop at the bottom and then twist it around in one direction many times, you will get our familiar double-stranded helix. Now, while keeping firm hold of the loop at the bottom, insert a pencil between the strands near the near the middle of the cord's length and then force the pencil downward. You will observe that the stands become progressively more tightly wound beneath your pencil, until it can move no more. At the same time the cord above the pencil becomes more loosely wound. Alternatively, you can let go the loop at the bottom, in which case the cord will spin around as the pencil descends.
This is relevant to the chromosome because when a gene is transcribed![]() , its two
double helical strands need to be separated, or "unzipped", as the
transcribing enzyme
, its two
double helical strands need to be separated, or "unzipped", as the
transcribing enzyme![]() moves along.
How, then, does the chromosome accommodate the twisting forces imposed by
this local "unzipping" of its two strands? You might expect the
chromosome to spin like the cord with a pencil moving down it. Certainly
there is some such movement. But if it were to proceed in an
unconstrained manner, as with the released window shade cord, the entire
chromosome ahead of the transcribing enzyme
moves along.
How, then, does the chromosome accommodate the twisting forces imposed by
this local "unzipping" of its two strands? You might expect the
chromosome to spin like the cord with a pencil moving down it. Certainly
there is some such movement. But if it were to proceed in an
unconstrained manner, as with the released window shade cord, the entire
chromosome ahead of the transcribing enzyme![]() would have
to make about 2850 complete turns during transcription of an average-sized
gene (Lavelle 2009), which means rotating at several turns per second.
Clearly, given the length, the mass, and the complex bending and looping
forms of the chromosome, and given the extremely thick "soup" of
macromolecules in the cell nucleus, such movement would be greatly
impeded.
would have
to make about 2850 complete turns during transcription of an average-sized
gene (Lavelle 2009), which means rotating at several turns per second.
Clearly, given the length, the mass, and the complex bending and looping
forms of the chromosome, and given the extremely thick "soup" of
macromolecules in the cell nucleus, such movement would be greatly
impeded.
Furthermore, the ends and many points within the chromosome are typically
"fastened down", as we will see later, so there isn't all that much
freedom of movement. Of course, when you move the pencil down between the
two strands of the window cord, you could allow the pencil itself to spin.
This would leave the helical structure of the cord mostly unchanged
outside the immediate vicinity of the pencil. However, in the cell our
"pencil" — the transcribing enzyme — is part of a very large
molecular complex. In addition, it is associated with a cumbersome set of
proteins for disassembling nucleosomes![]() ahead of its
transcribing activity, reassembling them behind, and performing various
other tasks. And it is attached to the ever-lengthening strand of RNA
ahead of its
transcribing activity, reassembling them behind, and performing various
other tasks. And it is attached to the ever-lengthening strand of RNA![]() that it is itself producing. So it
faces limits upon its mobility similar to those of the chromosome.
that it is itself producing. So it
faces limits upon its mobility similar to those of the chromosome.
The upshot of it all is that there are many complex movements, highly
constrained and absorbed in varying ways by the different resistant
elements of the complex structures involved. In general, positive
supercoiling![]() occurs ahead
of the transcribing enzyme's "unzipping" action and negative supercoiling
behind it. Topoisomerases play their role in managing both the stresses
and the overall conformation of the chromosome "tangle", as do many other
poorly characterized players in the sculptural drama of form and force
that is the chromosome.
occurs ahead
of the transcribing enzyme's "unzipping" action and negative supercoiling
behind it. Topoisomerases play their role in managing both the stresses
and the overall conformation of the chromosome "tangle", as do many other
poorly characterized players in the sculptural drama of form and force
that is the chromosome.
I have so far described the packaging of human DNA![]() as a mere technical challenge. That's a
big problem. The tensions and movements, the bending and unbending, the
coiling and uncoiling, are much more than the expression of mechanical
forces aimed at chromosome
as a mere technical challenge. That's a
big problem. The tensions and movements, the bending and unbending, the
coiling and uncoiling, are much more than the expression of mechanical
forces aimed at chromosome![]() condensation. It was quite wrong of me to begin by asking you to imagine
twisting a rope since, after all, there is no one — no specific
agent — in the cell nucleus performing this task. The chromosome is
not a passive, limp object moved only from outside. Interacting with its
surroundings, it is as much a living actor as any other part of its living
environment. Maybe instead of a rope, we should think of a snake,
coiling, curling, and sliding over a landscape that is itself in continual
movement.
condensation. It was quite wrong of me to begin by asking you to imagine
twisting a rope since, after all, there is no one — no specific
agent — in the cell nucleus performing this task. The chromosome is
not a passive, limp object moved only from outside. Interacting with its
surroundings, it is as much a living actor as any other part of its living
environment. Maybe instead of a rope, we should think of a snake,
coiling, curling, and sliding over a landscape that is itself in continual
movement.
The chromosome, in other words, is doing something. It is engaged in a highly effective spatial performance. It's movements are not simply the result of its being packaged and kept out of trouble, but rather are well-shaped responses to sensitively discerned needs. These movements bear decisive significance for the life of the cell and organism as a whole. Far better to picture the chromosome as both a sensing and muscular presence than as a rope.
To begin with, the mechanical stresses induced by transcription![]() are now
known to contribute broadly to gene regulation
are now
known to contribute broadly to gene regulation![]() . "The
organization of global transcription is tightly coupled to distribution of
supercoiling
. "The
organization of global transcription is tightly coupled to distribution of
supercoiling![]() sensitivity
in the genome"
sensitivity
in the genome"![]() (Blot 2006).
Increases in twist (positive supercoiling) are associated with chromatin
(Blot 2006).
Increases in twist (positive supercoiling) are associated with chromatin![]() folding and
gene silencing
folding and
gene silencing![]() in the
supercoiled region, whereas decreases of twist (negative supercoiling) are
associated with "acquisition of transcriptional competence" (Travers and
Muskhelishvili 2006). Moreover, "negative supercoils are dynamic. The
slithering and branching
in the
supercoiled region, whereas decreases of twist (negative supercoiling) are
associated with "acquisition of transcriptional competence" (Travers and
Muskhelishvili 2006). Moreover, "negative supercoils are dynamic. The
slithering and branching![]() of the
interwound strands allow DNA
of the
interwound strands allow DNA![]() to act like a chaperone
to act like a chaperone![]() , promoting
the long-range assembly and disassembly of protein-DNA complexes" (Deng et
al. 2005) — complexes that play a vital role in gene regulation.
Each type of cell has its own characteristic patterns of supercoiling,
which is doubtless related to the fact that it also has its own
distinctive patterns of gene expression
, promoting
the long-range assembly and disassembly of protein-DNA complexes" (Deng et
al. 2005) — complexes that play a vital role in gene regulation.
Each type of cell has its own characteristic patterns of supercoiling,
which is doubtless related to the fact that it also has its own
distinctive patterns of gene expression![]() . Christophe
Lavelle of the Curie Institute in France summarizes the recent research
findings this way:
. Christophe
Lavelle of the Curie Institute in France summarizes the recent research
findings this way:
As DNA is rotating inside the polymerase[transcribing enzyme], positive and negative supercoiling is induced downstream and upstream
, respectively. Transcriptionally generated torsion, rather than a mere waste product to be disposed of by topoisomerases, has instead recently been shown to propagate through the chromatin fiber and trigger local DNA alterations, detected as a regulatory signal by molecular partners. (Lavelle 2009)
To illustrate the regulatory possibilities: researchers at the National
Cancer Institute in Bethesda, Maryland, found that negative supercoiling![]() upstream
(behind) a transcribing enzyme was sufficient to cause a local,
nonstandard conformation of the double helix
upstream
(behind) a transcribing enzyme was sufficient to cause a local,
nonstandard conformation of the double helix![]() , which in
turn enabled recruitment of regulatory proteins sensitive to such changes
in structure (Kouzine et al. 2008).
, which in
turn enabled recruitment of regulatory proteins sensitive to such changes
in structure (Kouzine et al. 2008).
So the chromosome's twisting and writhing is not merely arbitrary; it is sculpturally significant movement, carrying meaning for the chromosomal stretches along which it is communicated.
But there are many other dimensions of the chromosome's![]() spatial
performance. Each chromosome has its own preferred territory within the
nucleus and its preferred neighbors, which also differ from one cell type
to another. These territories "are dynamic and plastic structures" that
"can be dynamically repositioned" (Schneider and Grosschedl 2007). Since
living conditions are close, the neighbors matter. A chromosome's
territory appears to be shaped rather like an irregular potato or a
sponge. There is at least some socializing between adjacent chromosomes,
with protrusions of one territory penetrating into the hollowed-out
portions of the next territory and even of more remote territories (Ling
et al. 2007). So not only are distantly separated portions of the same
chromosome brought into intimate contact by the geometry of the sponges,
but loci on separate chromosomes can also be brought into contact.
spatial
performance. Each chromosome has its own preferred territory within the
nucleus and its preferred neighbors, which also differ from one cell type
to another. These territories "are dynamic and plastic structures" that
"can be dynamically repositioned" (Schneider and Grosschedl 2007). Since
living conditions are close, the neighbors matter. A chromosome's
territory appears to be shaped rather like an irregular potato or a
sponge. There is at least some socializing between adjacent chromosomes,
with protrusions of one territory penetrating into the hollowed-out
portions of the next territory and even of more remote territories (Ling
et al. 2007). So not only are distantly separated portions of the same
chromosome brought into intimate contact by the geometry of the sponges,
but loci on separate chromosomes can also be brought into contact.
It happens that both sorts of contact have a great deal to do with gene
expression![]() . On an
earlier view, the DNA sequences
. On an
earlier view, the DNA sequences![]() regulating a
protein-coding gene were always close to the gene or at least not very far
removed. But in more recent years it's been recognized that some
regulatory sites — "enhancers"
regulating a
protein-coding gene were always close to the gene or at least not very far
removed. But in more recent years it's been recognized that some
regulatory sites — "enhancers"![]() and
"silencers"
and
"silencers"![]() and "locus
control regions"
and "locus
control regions"![]() — may
be located on distant parts of the chromosome, thousands or hundreds of
thousands or even millions of base pairs
— may
be located on distant parts of the chromosome, thousands or hundreds of
thousands or even millions of base pairs![]() away from
the gene being regulated
away from
the gene being regulated![]() . Expression
is enabled, for example, when the distant enhancer is brought into
physical proximity with the gene or genes it regulates. (Another
remarkable feat of contextually apt physical coordination!) In connection
with this, an activator
. Expression
is enabled, for example, when the distant enhancer is brought into
physical proximity with the gene or genes it regulates. (Another
remarkable feat of contextually apt physical coordination!) In connection
with this, an activator![]() protein is
bound to the enhancer and then, perhaps in concert with one or more
co-activators
protein is
bound to the enhancer and then, perhaps in concert with one or more
co-activators![]() , may assist
in constellating the massive transcription complex
, may assist
in constellating the massive transcription complex![]() on the
gene's promoter
on the
gene's promoter![]() sequence
sequence![]() .
.
A locus control region![]() (LCR) is a
DNA
(LCR) is a
DNA![]() sequence
that helps to regulate a cluster of related genes. One research team in
the Netherlands, working with mice, examined an LCR for a set of genes
relating to the production of beta-globin (a constituent of hemoglobin).
In fetal liver tissue, where these genes are highly active, the LCR was
found to associate with dozens of genes, including many involved in
beta-globin production. Some of these genes were tens of millions of base
pairs
sequence
that helps to regulate a cluster of related genes. One research team in
the Netherlands, working with mice, examined an LCR for a set of genes
relating to the production of beta-globin (a constituent of hemoglobin).
In fetal liver tissue, where these genes are highly active, the LCR was
found to associate with dozens of genes, including many involved in
beta-globin production. Some of these genes were tens of millions of base
pairs![]() distant on
the chromosome. Further, in fetal brain tissue, where the beta-globin
genes are inactive, the LCR again associated with many other sites —
but now a completely different set. The researchers concluded:
distant on
the chromosome. Further, in fetal brain tissue, where the beta-globin
genes are inactive, the LCR again associated with many other sites —
but now a completely different set. The researchers concluded:
Our observations demonstrate that not only active, but also inactive, genomic regions can transiently interact over large distances with many loci in the nuclear space. The data strongly suggest that each DNA segment has its own preferred set of interactions. This implies that it is impossible to predict the long-range interaction partners of a given DNA locus without knowing the characteristics of its neighboring segments and, by extrapolation, the whole chromosome. (Simonis et al. 2006)
It's not only where loci on a single chromosome![]() are brought
into contact with each other that they can interact, however. Increasing
numbers of cases are being reported where contact between sites on
different chromosomes plays a crucial role in gene regulation
are brought
into contact with each other that they can interact, however. Increasing
numbers of cases are being reported where contact between sites on
different chromosomes plays a crucial role in gene regulation![]() . In fact,
the mouse study just cited demonstrated a number of such contacts. (A few
researchers began to speak of "kissing chromosomes" — a not very
helpful phrase that seems now to have been dropped from the literature.)
Here's a schematic representation of one case of interchromosomal
interaction. (Skip the next three paragraphs if you don't want to bother
with the technical details.)
. In fact,
the mouse study just cited demonstrated a number of such contacts. (A few
researchers began to speak of "kissing chromosomes" — a not very
helpful phrase that seems now to have been dropped from the literature.)
Here's a schematic representation of one case of interchromosomal
interaction. (Skip the next three paragraphs if you don't want to bother
with the technical details.)
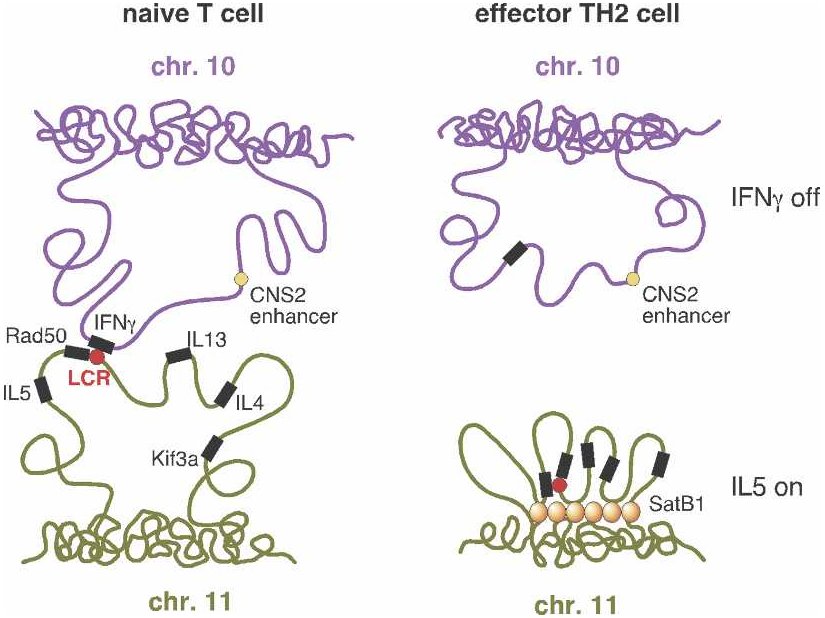 From Schneider and Grosschedl 2007. |
"T helper" or TH cells are human immune system cells. One type of TH cell (TH1) produces, among other things, interferon (IFN-gamma), while a second type (TH2) produces various interleukins such as IL-4 and IL-5. But before a cell becomes either a TH1 or TH2, it resides in a less differentiated state as a "naive" TH cell (referred to as a "T cell" in the figure). Only after stimulation by an antigen (a substance that provokes formation of antibodies), does the TH cell become either a TH1 or TH2 cell.
If you look at the genes responsible for producing interferon in TH1 and
interleukins in TH2 cells, you find the usual suspects: the interferon
gene on chromosome 10 seems to be regulated by nearby genomic elements,
while the interleukin genes on chromosome 11 are also regulated by local
sites — in particular, by an LCR![]() . But a research team at the Yale
University School of Medicine decided to investigate the larger spatial
picture. They discovered that regulatory regions
. But a research team at the Yale
University School of Medicine decided to investigate the larger spatial
picture. They discovered that regulatory regions![]() associated
with the interferon and interleukin genes, despite being on separate
chromosomes, were physically close together in naive cells. Upon
stimulation of naive cells by an antigen, the "negotiations" between these
regulatory regions somehow determined which of the genes would be active
and which would be repressed — and therefore whether the cell would
become a TH1 or TH2 cell. Following this determination, the two
chromosome regions moved apart.
associated
with the interferon and interleukin genes, despite being on separate
chromosomes, were physically close together in naive cells. Upon
stimulation of naive cells by an antigen, the "negotiations" between these
regulatory regions somehow determined which of the genes would be active
and which would be repressed — and therefore whether the cell would
become a TH1 or TH2 cell. Following this determination, the two
chromosome regions moved apart.
The illustration (right side) shows the case where a TH2 cell has
resulted. Interferon (IFN-gamma) is not expressed![]() (upper
right) because the looping pattern separates the requisite gene from its
enhancer
(upper
right) because the looping pattern separates the requisite gene from its
enhancer![]() . On the
other hand, IL-5 is expressed (lower right), because the protein
SATB1, which plays a large role in chromatin
. On the
other hand, IL-5 is expressed (lower right), because the protein
SATB1, which plays a large role in chromatin![]() organization
and transcription
organization
and transcription![]() regulation,
has anchored a series of chromosome loops in just such a way as to bring
the IL-5 gene and Rad50 promoter
regulation,
has anchored a series of chromosome loops in just such a way as to bring
the IL-5 gene and Rad50 promoter![]() into
proximity with the locus control region. (Spilianakis et al. 2005; see
also commentary in Kioussis 2005.)
into
proximity with the locus control region. (Spilianakis et al. 2005; see
also commentary in Kioussis 2005.)
Of course, as researchers dealing with this sort of thing readily acknowledge, questions abound. What guides particular sites on two different chromosomes to their rendezvous, and what sees to their subsequent separation? One could imagine, in the case of distantly separated sites on the same chromosome, that a regulatory protein binds to the one locus and then "tracks" along the chromosome until it finds the second locus (which it must have some way to recognize as significantly related to the first). But it's not at all easy to picture what it is that selects and brings together many loci on different chromosomes.
As is evident from the case of TH cells, chromosome looping can keep sites
apart as well as bring them together. It not only serves the purpose of
expression![]() , but also of
repression
, but also of
repression![]() . In a study
of red blood cells, a group of scientists from Children's Hospital in
Philadelphia showed that successive stages of cell maturation were marked
by different proteins playing a direct role in reconfiguring chromosome
loops — first for expression of a particular gene, and later for
repression (Jing et al. 2008). In general, chromosome loops help to make
possible the more or less independent regulation of different gene regions
— an important role in an environment thick with diverse regulatory
factors and processes.
. In a study
of red blood cells, a group of scientists from Children's Hospital in
Philadelphia showed that successive stages of cell maturation were marked
by different proteins playing a direct role in reconfiguring chromosome
loops — first for expression of a particular gene, and later for
repression (Jing et al. 2008). In general, chromosome loops help to make
possible the more or less independent regulation of different gene regions
— an important role in an environment thick with diverse regulatory
factors and processes.
But how does a locus on a chromosome![]() "take off"
through the three-dimensional space of the nucleus, uncoiling from a more
condensed state into a thin thread and looping outward from its territory
for considerable distances, as if drawn by an invisible hand toward a
rendezvous with a distant location? One group of researchers positioned a
transcriptional activator
"take off"
through the three-dimensional space of the nucleus, uncoiling from a more
condensed state into a thin thread and looping outward from its territory
for considerable distances, as if drawn by an invisible hand toward a
rendezvous with a distant location? One group of researchers positioned a
transcriptional activator![]() on a
particular chromosomal site located close to the periphery of the cell
nucleus. Within 1 - 2 hours, the site migrated to the interior of the
nucleus, following a curvilinear path roughly perpendicular to the nuclear
envelope
on a
particular chromosomal site located close to the periphery of the cell
nucleus. Within 1 - 2 hours, the site migrated to the interior of the
nucleus, following a curvilinear path roughly perpendicular to the nuclear
envelope![]() . The
movement, which was interspersed with several-minute periods of
quiescence, reached a maximum velocity of about 1/10 the nuclear diameter
per minute. These results led the researchers to speak of "fast and
directed long-range chromosome movements" (Chuang et al. 2006).
. The
movement, which was interspersed with several-minute periods of
quiescence, reached a maximum velocity of about 1/10 the nuclear diameter
per minute. These results led the researchers to speak of "fast and
directed long-range chromosome movements" (Chuang et al. 2006).
A fairly recent surprise has been the discovery of actin and myosin in connection with some chromosome movements. These two substances, which play a major role in the contraction of muscles, seem also to provide a kind of "musculature" within the nucleus. Get rid of them, and certain observed movements stop. But little is yet known about how the movement is actually achieved, and even less about how it is directed.
What is now known, however, is that the nucleus is much more than a
linear assembly line for the construction of proteins based on genetic
sequences![]() . It
participates in an elaborately organized, three-dimensional space, and the
positioning and movement of both chromosomes and the regulatory elements
within the nucleus have everything to do with the functioning of the
genome
. It
participates in an elaborately organized, three-dimensional space, and the
positioning and movement of both chromosomes and the regulatory elements
within the nucleus have everything to do with the functioning of the
genome![]() .
.
But while extraordinary research energies are now directed toward
articulating the undeniable structural organization of the cell nucleus in
its relation to gene regulation![]() , an overall,
coherent picture of the organization remains elusive. The titles of
several articles currently lying on my desk point to the challenge
investigators face:
, an overall,
coherent picture of the organization remains elusive. The titles of
several articles currently lying on my desk point to the challenge
investigators face:
"Dynamics and Interplay of Nuclear Architecture, Genome Organization, and Gene Expression" (Schneider and Grosschedl 2007)."Dynamic Genome Architecture in the Nuclear Space: Regulation of Gene Expression in Three Dimensions" (Lanctôt et al. 2007).
"The Third Dimension of Gene Regulation: Organization of Dynamic Chromatin Loopscape by SATB1" (Galande et al. 2007).
"Dynamic Regulation of Nucleosome Positioning in the Human Genome" (Schones et al. 2008).
"Dynamic Organization of Gene Loci and Transcription Compartments in the Cell Nucleus" (Spudich 2008).
"Nuclear Functions in Space and Time: Gene Expression in a Dynamic, Constrained Environment" (Trinkle-Mulcahy and Lamond 2008).
You will have noted the repeated juxtaposition — spatial
organization on the one hand, dynamism on the other. How does one capture
organization that is dynamic and ever-shifting? The question only becomes
more acute when we look at a few additional aspects of nuclear
organization, as currently described in the literature:
Chromosome Domains. Chromosomes![]() , as we have
seen, participate in the highly structured space of the nucleus. But that
is not all. They themselves are structured along their length, being
subdivided by various means and in ever-changing ways into chromosome
domains. We've already seen the organization of the chromosome into
densely compacted regions (known as heterochromatin
, as we have
seen, participate in the highly structured space of the nucleus. But that
is not all. They themselves are structured along their length, being
subdivided by various means and in ever-changing ways into chromosome
domains. We've already seen the organization of the chromosome into
densely compacted regions (known as heterochromatin![]() ) and less
condensed, more active regions (euchromatin
) and less
condensed, more active regions (euchromatin![]() ). The
boundaries between such regions are not always well-defined. Simply by
residing close to a region of heterochromatin, a gene that otherwise would
be very actively transcribed
). The
boundaries between such regions are not always well-defined. Simply by
residing close to a region of heterochromatin, a gene that otherwise would
be very actively transcribed![]() might be
only intermittently expressed
might be
only intermittently expressed![]() , or even
silenced
, or even
silenced![]() altogether.
Where somewhat more cleanly separate regulation of neighboring loci is
important, special DNA sequences
altogether.
Where somewhat more cleanly separate regulation of neighboring loci is
important, special DNA sequences![]() called
insulators
called
insulators![]() can help
prevent the "leaking" of influence from one region of the chromosome to
the next.
can help
prevent the "leaking" of influence from one region of the chromosome to
the next.
Chromosome domains are also established by the twisting forces (torsion)
communicated more or less freely along bounded segments of the chromosome.
(The boundaries might be defined, for example, by the tethering points of
chromosome loops.) The loci within such a region share a common torsion,
and this can attract a common set of regulatory proteins. The torsion
also tends to correlate with the level of compaction of the chromatin
fiber, which in turn correlates with many other aspects of gene
regulation![]() . And even
on an extremely small scale, the twisting (by linker histones
. And even
on an extremely small scale, the twisting (by linker histones![]() ) of the
short stretches of DNA between nucleosomes
) of the
short stretches of DNA between nucleosomes![]() — or
the untwisting brought about by the release of the histones — is
presumed to drive the folding or unfolding of the local chromatin
— or
the untwisting brought about by the release of the histones — is
presumed to drive the folding or unfolding of the local chromatin![]() (Travers and
Muskhelishvili 2006). All this reminds us that gene regulation is defined
less by static entities than by the quality and force of various
movements.
(Travers and
Muskhelishvili 2006). All this reminds us that gene regulation is defined
less by static entities than by the quality and force of various
movements.
There are still other ways that the chromosome reveals itself as a dynamic, complexly structured context. Genes expressed in the same cell type or at the same time, genes sharing common regulatory factors, and genes actively expressed (or mostly inactive) tend to be grouped together. One way such domains could be established is through the binding of the same protein complexes along a region of the chromosome, thereby establishing a common molecular and regulatory environment for the encompassed genes. But it's important to realize that such regions are more a matter of tendency than of absolute rule. A few examples from a summary by Elzo de Wit and Bas van Steensel at the Netherlands Cancer Institute illustrate the situation:
These rough tendencies do not enable precise predictions, but yet the
tendencies are really there; they point toward meaningful organization,
even though the observed "rules" are less the determiners of that
organization than they are the continually modified products of it. This
is exactly what you would expect in any living context, where a larger
unity — the unity that leads us to refer quite naturally to a
living being — shapes the activity of local parts and
processes to its own intention.
The "Pull" of the Nuclear
Periphery. There is a fibrous network — the "nuclear lamina"
— located primarily at the inside face of the nuclear envelope.
In vitro studies (what used to be referred to as "test-tube
studies") have shown that several proteins of this network can interact
with chromatin![]() . And now,
as the Dutch biologists summarize, it has been found that there are more
than 1300 lamina-associated chromosome domains (LADs) in the human
genome
. And now,
as the Dutch biologists summarize, it has been found that there are more
than 1300 lamina-associated chromosome domains (LADs) in the human
genome![]() that do
indeed preferentially locate themselves at the nuclear periphery. De Wit
and van Steensel (2009) mention studies showing that the artificial
anchoring of a chromosome
that do
indeed preferentially locate themselves at the nuclear periphery. De Wit
and van Steensel (2009) mention studies showing that the artificial
anchoring of a chromosome![]() locus to the
nuclear lamina "can cause partial downregulation of some (but not all)
genes surrounding the anchoring sequence
locus to the
nuclear lamina "can cause partial downregulation of some (but not all)
genes surrounding the anchoring sequence![]() ".
".
There seems to be a general rule that the chromosomes and chromosome
territories located toward the periphery of the nucleus are less
transcriptionally![]() active and
also less gene-dense. Conversely, the nuclear interior shows higher rates
of activity and greater gene density. Researchers can activate
active and
also less gene-dense. Conversely, the nuclear interior shows higher rates
of activity and greater gene density. Researchers can activate![]() genes near
the nuclear envelope and then watch them as they move toward the interior.
Likewise, they can silence
genes near
the nuclear envelope and then watch them as they move toward the interior.
Likewise, they can silence![]() genes in the
interior and watch them relocating to the periphery. Nevertheless,
transcription does occur in the outlying regions, and silent regions of
chromosomes reside in the interior. And, as always, multiple dimensions
of regulation work together. For example, the radial positioning of genes
seems to be connected to specific histone modifications
genes in the
interior and watch them relocating to the periphery. Nevertheless,
transcription does occur in the outlying regions, and silent regions of
chromosomes reside in the interior. And, as always, multiple dimensions
of regulation work together. For example, the radial positioning of genes
seems to be connected to specific histone modifications![]() of the sort
we looked at in Part 1 — although it's a matter of rough rather than
absolutely consistent correlation (Strašák et al. 2009).
of the sort
we looked at in Part 1 — although it's a matter of rough rather than
absolutely consistent correlation (Strašák et al. 2009).
Nuclear Matrix. It is not only the peripheral nuclear lamina that
provides a kind of skeletal structure for organizing the nucleus. There
is, throughout the nuclear space, a still poorly characterized and elusive
"nuclear matrix" — so elusive that its fundamental nature is still
debated. The nuclear lamina![]() can be
considered part of it, and there are many other substances that seem to
play a role, including the SATB1 protein we encountered in connection with
looping chromosomes, a topoisomerase
can be
considered part of it, and there are many other substances that seem to
play a role, including the SATB1 protein we encountered in connection with
looping chromosomes, a topoisomerase![]() , actin
, actin![]() , and even
the DNA transcribing
, and even
the DNA transcribing![]() and
replication
and
replication![]() enzymes.
Many of the proteins that associate with chromatin
enzymes.
Many of the proteins that associate with chromatin![]() , affecting
its form and compaction, are considered to be components of the nuclear
matrix. In other words, the nuclear matrix is not simply a passive
structure that objects can attach to. It consists of active agents
— and, in our current context, that means agents of gene
regulation
, affecting
its form and compaction, are considered to be components of the nuclear
matrix. In other words, the nuclear matrix is not simply a passive
structure that objects can attach to. It consists of active agents
— and, in our current context, that means agents of gene
regulation![]() .
.
The human genome![]() contains an
estimated 30,000-80,000 "matrix attachment regions" (MARs) —
relatively short DNA sequences susceptible of being anchored to the
nuclear matrix (Ottaviani et al. 2008). These anchoring points can
contribute to the formation of the loops we've been talking about. Some
MARs are more or less permanently attached to the matrix and may be
associated with higher-order chromatin
contains an
estimated 30,000-80,000 "matrix attachment regions" (MARs) —
relatively short DNA sequences susceptible of being anchored to the
nuclear matrix (Ottaviani et al. 2008). These anchoring points can
contribute to the formation of the loops we've been talking about. Some
MARs are more or less permanently attached to the matrix and may be
associated with higher-order chromatin![]() compaction
and the repression
compaction
and the repression![]() of genes not
required in a particular cell type. Others only transiently attach to the
matrix and are thought to play a major role in the management of gene
expression
of genes not
required in a particular cell type. Others only transiently attach to the
matrix and are thought to play a major role in the management of gene
expression![]() . The
configuration of attachments at any given moment shapes the overall
chromosome architecture, and the consequent looping patterns effectively
insulate some regions from regulatory factors while exposing others. In
sum:
. The
configuration of attachments at any given moment shapes the overall
chromosome architecture, and the consequent looping patterns effectively
insulate some regions from regulatory factors while exposing others. In
sum:
Our understanding of how the genome functions in the context of the nucleus has been propelled by indisputable evidence that distinct genomic sites bind to regulatory proteins at the nuclear matrix. The emerging picture is that these genomic anchors regulate transcriptionand replication
by dynamically organizing chromatin in three-dimensional space. (Ottaviani et al. 2008)
By this time I'm sure you recognize the need to ask (upon hearing that
genomic anchors "regulate transcription"): What is it that regulates the
anchors? How do they know when and where and for how long to participate
in their anchoring task? The point, which is one of our enduring themes
in these articles, is that there is no single, controlling level of gene
regulation, subordinating the rest of the organism to itself. Noting that
"the existence of regulatory cross-talk between spatially interacting loci
opens up a new dimension in the study of gene regulation", Christian
Lanctôt et al. (2007) go on to remark that "not only does [this
cross-talk] constitute an additional level of complexity in the search for
regulatory elements in the genome![]() , it also
implies that chromatin
, it also
implies that chromatin![]() mobility
itself, and therefore the ensuing long-range gene-gene interactions, might
be a target of regulation".
mobility
itself, and therefore the ensuing long-range gene-gene interactions, might
be a target of regulation".
How could it be otherwise within a harmoniously functioning organism? I
suspect it's quite safe to say that every aspect of the cell is in one way
or another a target of regulation, and at the same time takes on some of
the role of regulator. Or better (since that last statement reduces the
word "regulation" to something close to nonsense): every part participates
in the whole organism and is informed by the whole. Of course, in the
current state of biology this remark, too, will strike many as nonsense.
One could reply by asking whether it's any more nonsensical than
all the usual talk of "regulation", but a more positive approach would be
to take up the question of holism, as we will do later in this series.
Nuclear Compartments and Organelles. Loops are created
when separate points on a chromosome![]() are brought
together and at least temporarily bound at the same location.
Multiple-loop structures result when a number of different loci fraternize
in this way. This raises the question: in what sense are the regions
where these gatherings occur "real places"? That is, what structural
identity do they possess beyond the fact that they happen to be sites
where active chromosomal loci have gathered?
are brought
together and at least temporarily bound at the same location.
Multiple-loop structures result when a number of different loci fraternize
in this way. This raises the question: in what sense are the regions
where these gatherings occur "real places"? That is, what structural
identity do they possess beyond the fact that they happen to be sites
where active chromosomal loci have gathered?
In one sense, the answer is easy. In order for genes to be active, there
must be transcribing enzymes![]() and many
other factors related to transcription
and many
other factors related to transcription![]() . So it
stands to reason that these centers of activity are distinctively
constituted. High-resolution surveys of the cell nucleus do in fact show
many such places, which have come to be called transcription "factories"
(a rather prejudicial term). Estimates for their number range from 500 to
10,000 (Trinkle-Mulcahy 2008), and it has been conjectured that, on
average, some eight transcribing enzymes are present in each center of
activity.
. So it
stands to reason that these centers of activity are distinctively
constituted. High-resolution surveys of the cell nucleus do in fact show
many such places, which have come to be called transcription "factories"
(a rather prejudicial term). Estimates for their number range from 500 to
10,000 (Trinkle-Mulcahy 2008), and it has been conjectured that, on
average, some eight transcribing enzymes are present in each center of
activity.
One group of researchers, describing how distantly separated genes in red
blood cells "colocalize to the same transcription factory at high
frequencies", go on to summarize the situation: "active genes are
dynamically organized into shared nuclear subcompartments and movement
into or out of these factories results in activation or abatement of
transcription. Thus, rather than recruiting and assembling transcription
complexes, active genes migrate to preassembled transcription sites"
(Osborne et al. 2004). The implication, noted by the authors, is that
"mechanisms regulating recruitment of genes into factories would be
expected to have a fundamental role in gene expression![]() ".
".
The transcribing enzymes![]() in (or,
rather, at the outer surface of) the active transcription centers,
according to the emerging view, do not themselves move along the genes;
they "reel in" the genes they are transcribing. In this way they act as
critical structural elements for maintaining the loops, which come and go
as the various enzymes and regulatory factors bind and release them
(Carter et al. 2008).
in (or,
rather, at the outer surface of) the active transcription centers,
according to the emerging view, do not themselves move along the genes;
they "reel in" the genes they are transcribing. In this way they act as
critical structural elements for maintaining the loops, which come and go
as the various enzymes and regulatory factors bind and release them
(Carter et al. 2008).
But, still, uncertainty remains about how much "there" is really there in
the transcription centers. To what degree do enduring structures exist
apart from the organized "structure" of the ongoing processes of
transcription? There is presumably something there, but it's
proven subtle and difficult to pin down. The matrix attachment regions![]() of chromosomes, which presumably play a
role in bringing genes to transcriptional centers, are being identified,
but it remains to find anything in the way of a very fixed and definite
structure for them to attach to. The "structure", such as it is, seems to
be as much process as product.
of chromosomes, which presumably play a
role in bringing genes to transcriptional centers, are being identified,
but it remains to find anything in the way of a very fixed and definite
structure for them to attach to. The "structure", such as it is, seems to
be as much process as product.
There are yet other nuclear compartments relating to gene expression, but we will not pursue them here.
The intricately formed activity of the nucleus varies from one cell type
to another and from one stage of an organism's development to another. It
both shapes and mirrors the distinctive character of the individual cell.
But this character is not some abstract essence detached from whatever
else is going on in the organism at a particular moment. We can only
assume that, whether the cat we are looking at is stalking or eating or
sleeping or raising its fur in a confrontation with an enemy, the
expressive differences we can recognize in these activities would be
matched by expressive differences at every level of the cat's life,
including the level of gene transcription![]() and nuclear
organization — if only we were capable of reading the cell with the
same qualitative attention we devote to the outward behavior of the cat.
If the cat is raising its fur, then the skin, muscle, and other cells must
in some sense be "raising their fur" as well.
and nuclear
organization — if only we were capable of reading the cell with the
same qualitative attention we devote to the outward behavior of the cat.
If the cat is raising its fur, then the skin, muscle, and other cells must
in some sense be "raising their fur" as well.
In other words, the chromosome![]() movements
we've looked at are always part of the larger activity of the organism.
It's not just that a locus of the chromosome moves from point A to point B
in order to connect with a group of other loci; this process in
turn takes place in order to achieve equally significant
performances at higher levels of observation, whether it's a matter of the
cell's response to a nearby lesion or to starvation or to the organism's
emotional state. The activities in the cell nucleus are part of an
overall organic picture, and the scientist will do well to remember
occasionally how remote is the detailed knowledge of the sort I've
outlined above from any coherent and contextual understanding of what's
going on.
movements
we've looked at are always part of the larger activity of the organism.
It's not just that a locus of the chromosome moves from point A to point B
in order to connect with a group of other loci; this process in
turn takes place in order to achieve equally significant
performances at higher levels of observation, whether it's a matter of the
cell's response to a nearby lesion or to starvation or to the organism's
emotional state. The activities in the cell nucleus are part of an
overall organic picture, and the scientist will do well to remember
occasionally how remote is the detailed knowledge of the sort I've
outlined above from any coherent and contextual understanding of what's
going on.
This is presumably why biologists Amy Hark and Steven Triezenberg (2001), speaking of the variety of protein complexes affecting chromatin structure and gene expression, point to "a web of functional interactions that might be viewed as either elegantly integrated or hopelessly tangled". Hopelessly tangled, that is, if we do no more than lose ourselves in tracking isolated "causal factors" and "effects"; elegantly integrated if we can somehow rise to a more pictorial and qualitative grasp of what clearly is in fact a unified whole.
Perhaps we have the most incentive to seek such a wider understanding when we confront disease. There is no doubt, for example, that the phenomena investigated by the epigeneticist bear heavily on cancer, even if there is little effort as yet to read the cell as an expressive whole. Certainly research into particular "mechanisms" is proceeding at full tilt. Referring to how the microenvironments of the nucleus bring together the various gene-regulatory signals in all their necessary combinations, one team of researchers reviews the implications for cancer diagnosis and treatment:
Solid tumours, leukaemias, and lymphomas show striking alterations in nuclear morphology as well as in the architectural organization of genes, transcriptsThe researchers add that the effects of therapeutic treatment hang in the balance of these complex interactions, since even a patient's sensitivity to radiation and chemotherapy depends on the "composition, assembly and architectural organization of regulatory machinery within the cancer cell nucleus". The hope, finally, is that the "functional relationship between nuclear organization and gene expression, and regulatory
complexes within the nucleus. . . . These cancer-related changes disrupt several levels of nuclear organization that include linear gene sequences, chromatin
organization and subnuclear [compartments]. . . . Modifications in chromatin remodelling complexes
, the persistent association of regulatory proteins with gene loci, and DNA methylation
epigenetically modulate genome
accessibility to regulatory factors for the physiological control of cell fate.... (Zaidi et al. 2007).
That hope may sooner or later be fulfilled. But the scale of the challenge looks hard to underestimate!
In any case, the fluid spatial organization of the nucleus and the
movement of chromosomes![]() within it
clearly play a vital role in bringing about the right "marriages" between
participants in the intricate playing out of genomic expression —
and also in avoiding the wrong marriages. The evidence suggests,
according to UK geneticists Peter Fraser and Wendy Bickmore, that "the
dynamic spatial organization of the nucleus both reflects and shapes
genome
within it
clearly play a vital role in bringing about the right "marriages" between
participants in the intricate playing out of genomic expression —
and also in avoiding the wrong marriages. The evidence suggests,
according to UK geneticists Peter Fraser and Wendy Bickmore, that "the
dynamic spatial organization of the nucleus both reflects and shapes
genome![]() function. . . . We now have a picture of a genome that
is 'structured', not in a rigid three-dimensional network, but in a
dynamic organization [that] clearly changes during normal development
function. . . . We now have a picture of a genome that
is 'structured', not in a rigid three-dimensional network, but in a
dynamic organization [that] clearly changes during normal development![]() and
differentiation"
and
differentiation"![]() (Fraser and
Bickmore 2007).
(Fraser and
Bickmore 2007).
We began by asking ourselves how the cell condenses two meters of DNA![]() into a nucleus ten millionths of a meter
in diameter. The question is justified, but we can see by now that it's
hardly a mere matter of avoiding a snarled state so that an autonomous
logic of transcription
into a nucleus ten millionths of a meter
in diameter. The question is justified, but we can see by now that it's
hardly a mere matter of avoiding a snarled state so that an autonomous
logic of transcription![]() can proceed
along its fated way. The adroit dynamics and deft sculpturing of
chromosomes and an entire galaxy of proteins are as much the "whole point
of the show" as any fixed code. The logic of transcription itself is, at
least in part, a disciplined art of movement. The next time you find
yourself picturing heredity as the transmission of fixed, determinative
elements from parent to offspring, you might pause to ask yourself how
such statically imagined elements could determine the art of movement that
also comes to expression in successive generations.
can proceed
along its fated way. The adroit dynamics and deft sculpturing of
chromosomes and an entire galaxy of proteins are as much the "whole point
of the show" as any fixed code. The logic of transcription itself is, at
least in part, a disciplined art of movement. The next time you find
yourself picturing heredity as the transmission of fixed, determinative
elements from parent to offspring, you might pause to ask yourself how
such statically imagined elements could determine the art of movement that
also comes to expression in successive generations.
There is, after all, as much cause and effect, as much determination of
outcomes, as much logic and reason, in the compaction and twisting,
the movement and re-shaping, as there is in any other aspect of the cell
nucleus, even if the dynamism is fluid and irreducible to digital terms.
The chromosome performs an unceasing dance and — crucially —
the ever-shifting pattern of the dance lends its form and organization
to the expression of genes. Perhaps that is why a pair of geneticists
could write — very wisely, I think — that trying to define the
chromatin![]() complex "is
like trying to define life itself" (Grewal and Elgin 2007).
complex "is
like trying to define life itself" (Grewal and Elgin 2007).
If we ignore the artful movement, it's not because we find in it little meaningful expression of the cell's nature, but only because we have a difficult time translating it into the familiar and preferred terms of science. But that's a limitation of our science, not of the cell. We already have enough evidence to say that the movement, as movement, must be at least as deft and graceful, and at least as well-calculated, as any Olympic gymnast's.
Do the genes control the cell and hand down instructions? Whatever reason there may be to view the matter from that angle, there's at least as much reason to think of the dance of chromosomes as controlling the genes. Which individual genes can be expressed and how much; which "signaling" functions of the cell are brought to bear on any particular gene; which large stretches of the chromosome are prepared for longer-term expression and which are put into "cold storage" — all this is not so much digitally enunciated as gestured by the entire context. And the choreography continually varies, summoning genes to participate in the power of its higher-order artistry.
It is not too much to say that the cell presents us with forms constantly modulated by the cellular environment and beyond — living sculptures, shape-shifting in response to a music we have not yet inquired about, let alone learned to hear.
(You will find the latest versions of the currently available parts of this series at the website, "From Mechanism to a Science of Qualities".)
Please Note: With a view toward the needs of the readership, I have preferred to cite review articles, where they are available and, in general, have made little effort to reflect in my citations the priority claims of the various investigators of any particular phenomenon. Public (online) accessibility of papers and ease of access to the relevant information are primary criteria for my selection — qualified, of course, by the limits of my own familiarity with the literature.
Blot, Nicolas, Ramesh Mavathur, Marcel Geertz, et al. (2006). "Homeostatic Regulation of Supercoiling Sensitivity Coordinates Transcription of the Bacterial Genome", EMBO Reports vol. 7, no. 7, pp. 710-5. doi:10.1038/sj.embor.7400729
Carter, David R. F., Christopher Eskiw, and Peter R. Cook (2008). "Transcription Factories", Biochemical Society Transactions vol. 36, pp. 585-9. doi:10.1042/BST0360585
Chuang, Chien-Hui, Anne E. Carpenter, Beata Fuchsova, et al. (2006). "Long-range Directional Movement of an Interphase Chromosome Site", Current Biology vol. 16 (Apr. 18), pp. 825-31. doi:10.1016/j.cub.2006.03.059
Costelloe, Thomas, Jennifer FitzGerald, Niall J. Murphy, et al. (2006). "Chromatin Modification and the DNA Damage Response", Experimental Cell Research vol. 312, pp. 2677-86. doi:10.1016/j.yexcr.2006.06.031
Deng, Shuang, Richard A. Stein, and N. Patrick Higgins (2005). "Organization of Supercoil Domains and Their Reorganization by Transcription", Molecular Microbiology vol. 57, no. 6 (September), pp. 1511-21. doi:10.1111/j.1365-2958.2005.04796.x
De Wit, Elzo and Bas van Steensel (2009). "Chromatin Domains in Higher Eukaryotes: Insights from Genome-wide Mapping Studies", Chromosoma vol. 118, pp. 25-36.
Fraser, Peter and Wendy Bickmore (2007). "Nuclear Organization of the Genome and the Potential for Gene Regulation", Nature vol. 447 (May 24), pp. 413-7. doi:10.1038/nature05916
Fraser, Peter and James Douglas Engel (2006). "Constricting Restricted Transcription: The (Actively?) Shrinking Web", Genes and Development vol. 20, pp. 1379-83. doi:10.1101/gad.1438106
Galande, Sanjeev, Prabhat Kumar Purbey, Dimple Notani, et al. (2007). "The Third Dimension of Gene Regulation: Organization of Dynamic Chromatin Loopscape by SATB1", Current Opinion in Genetics and Development vol. 17, pp. 408-14. doi:10.1016/j.gde.2007.08.003
Grewal, Shiv I. S. and Sarah C. R. Elgin (2007). "Transcription and RNA Interference in the Formation of Heterochromatin", Nature vol. 447 (May 24), pp. 399-406. doi:10.1038/nature05914
Hark, Amy T. and Steven J. Triezenberg (2001). "Chromatin and Transcription: Merging Package and Process" (review of Chromatin Structure and Gene Expression, edited by Sarah C. R. Elgin and Jerry L. Workman), Cell vol. 105, no. 3 (May 4), pp. 321-3. doi:10.1016/S0092-8674(01)00352-X
Jing, Huie, Christopher R. Vakoc, Lei Ying, et al. (2008). "Exchange of GATA Factors Mediates Transitions in Looped Chromatin Organization at a Developmentally Regulated Gene Locus", Molecular Cell vol. 29, no. 2 (Feb. 1), pp. 232-42. doi:10.1016/j.molcel.2007.11.020
Kioussis, Dimitris (2005). "Gene Regulation: Kissing Chromosomes", Nature vol. 435, no. 7049 (June 2), pp. 579-80. doi:10.1038/435579a
Kouzine, Fedor and David Levens (2007). "Supercoil-driven DNA Structures Regulate Genetic Transactions", Frontiers in Bioscience vol. 12 (May 1), pp. 4409-23).
Kouzine, Fedor, Suzanne Sanford, Zichrini Elisha-Feil, et al. (2008). "The Functional Response of Upstream DNA to Dynamic Supercoiling in Vivo", Nature Structural and Molecular Biology vol. 15, no. 2 (Feb.), pp. 146-54. doi:10.1038/nsmb.1372
Lanctôt, Christian, Thierry Cheutin, Marion Cremer, et al. (2007). "Dynamic Genome Architecture in the Nuclear Space: Regulation of Gene Expression in Three Dimensions", Nature Reviews Genetics vol. 8, no. 2 (Feb.), pp. 104-15. doi:10.1038/nrg2041
Lavelle, Christophe (2009). "Forces and Torques in the Nucleus: Chromatin under Mechanical Constraints", Biochemistry and Cell Biology vol. 87, pp. 307-22. doi:10.1139/O08-123
Ling, Jian Qun and Andrew R. Hoffman (2007). "Epigenetics of Long-Range Chromatin Interactions", Pediatric Research vol. 61, no. 5, Pt. 2, pp. 11R-16R. doi:10.1203/pdr.0b013e31804575db
Lior, David, Wolfgang Huber, Marina Granovskaia, et al. (2006). "A High-resolution Map of Transcription in the Yeast Genome", PNAS vol. 103, no. 14 (Apr. 4), pp. 5320-5. doi:10.1073/pnas.0601091103 http://www.pnas.org/content/103/14/5320.full.
Lomvardas, Stavros, Gilad Barnea, David J. Pisapia, et al. (2006). "Interchromosomal Interactions and Olfactory Receptor Choice", Cell vol. 126 (July 28), pp. 403-13. doi:10.1016/j.cell.2006.06.035
Misteli, Tom (2007). "Beyond the Sequence: Cellular Organization of Genome Function", Cell vol. 128 (Feb. 23), pp. 787-800. doi:10.1016/j.cell.2007.01.028
Osborne, Cameron S., Lyubomira Chakalova, Karen E. Brown, et al. (2004). "Active Genes Dynamically Colocalize to Shared Sites of Ongoing Transcription", Nature Genetics vol. 36, no. 10 (Oct.), pp. 1065-71. doi:10.1038/ng1423
Ottaviani, Diego, Elliott Lever, Petros Takousis, et al. (2008). "Anchoring the Genome", Genome Biology vol. 9, article 201 (Jan. 22). doi:10.1186/gb-2008-9-1-201
Schneider, Robert and Rudolf Grosschedl (2007). "Dynamics and Interplay of Nuclear Architecture, Genome Organization, and Gene Expression", Genes and Development vol. 21, pp. 3027-3043. doi:10.1101/gad.1604607
Schones, Dustin E., Kairong Cui, Suresh Cuddapah, et al. (2008). "Dynamic Regulation of Nucleosome Positioning in the Human Genome", Cell vol. 132 (Mar. 7), pp. 887-98. doi:10.1016/j.cell.2008.02.022
Simonis, Marieke, Petra Klous, Erik Splinter, et al. (2006). "Nuclear Organization of Active and Inactive Chromatin Domains Uncovered by Chromosome Conformation Capture-on-chip (4C)", Nature Genetics vol. 38, no. 11 (Nov.), pp. 1348-54. doi:10.1038/ng1896
Spilianakis, Charalampos G., Maria D. Lalioti, Terrence Town, et al. (2005). "Interchromosomal Associations between Alternatively Expressed Loci", Nature vol. 435, no. 7042 (June 2), pp. 637-45. doi:10.1038/nature03574
Spudich, James A. (2008). "Dynamic Organization of Gene Loci and Transcription Compartments in the Cell Nucleus", Biophysical Journal vol. 95, no. 11 (Dec. 1), pp. 5003-4. doi:10.1529/biophysj.108.139196
Strašák, Luděk, Eva Bártová, Andrea Harničarova, et al. (2009). "H3K9 Acetylation and Radial Chromatin Positioning", Journal of Cellular Physiology. doi:10.1002/jcp.21734. No other citation information provided by publisher.
Travers, Andrew and Georgi Muskhelishvili (2007). "A Common Topology for Bacterial and Eukaryotic Transcription Initiation?" EMBO Reports vol. 8, no. 2, pp. 147-51. doi:10.1038/sj.embor.7400898
Trinkle-Mulcahy, Laura and Angus I. Lamond (2008). "Nuclear Functions in Space and Time: Gene Expression in a Dynamic, Constrained Environment", FEBS Letters. doi:10.1016/j.febslet.2008.04.029
Zaidi, Sayyed K., Daniel W. Young, Amjad Javed, et al. (2007). "Nuclear Microenvironments in Biological Control and Cancer", Nature Reviews Cancer vol. 7 (June), pp. 454-63. doi:10.1038/nrc2149
NetFuture, a freely distributed electronic newsletter, is published and copyrighted by The Nature Institute. The editor is Stephen L. Talbott, author of Devices of the Soul: Battling for Our Selves in the Age of Machines (http://natureinstitute.org/txt/st). You may redistribute this newsletter for noncommercial purposes. You may also redistribute individual articles in their entirety, provided the NetFuture url and this paragraph are attached.
NetFuture is supported by freely given reader contributions, and could not survive without them. For details and special offers, see http://netfuture.org/support.html .
Current and past issues of NetFuture are available on the Web:
http://netfuture.org
To subscribe or unsubscribe, go to
http://netfuture.org/subscribe.html.If you have problems subscribing or unsubscribing, send mail to: owner-netfuture@lists.panix.com.
This issue of NetFuture: http://netfuture.org/2009/May2809_176.html.
Steve Talbott :: NetFuture #176 :: May 28, 2009