Two brief recommendations:
SLT
(You will find the latest versions of the currently available parts of this series at the website, "From Mechanism to a Science of Qualities".)
By clicking on the shaded rectangles at the end of many scientific terms, you can immediately read a definition of the terms in a separate window. This requires JavaScript to be enabled in your browser.
If you are like me, then upon hearing that the DNA![]() double helix
double helix![]() wraps a
couple of times around a histone
wraps a
couple of times around a histone![]() "spool" (of
which there are maybe 30 million in the human genome) and then, extending
a short distance beyond, wraps around the next spool, your imagination's
first response would probably be to summon a picture of more or less
smoothly machined cylinders, each one uniformly encircled by DNA. And, in
fact, the schematic illustrations one encounters in the literature do
sometimes represent the histone-DNA complex (called a "nucleosome") in
that way, as in the following drawing of three nucleosomes and various
associated molecules:
"spool" (of
which there are maybe 30 million in the human genome) and then, extending
a short distance beyond, wraps around the next spool, your imagination's
first response would probably be to summon a picture of more or less
smoothly machined cylinders, each one uniformly encircled by DNA. And, in
fact, the schematic illustrations one encounters in the literature do
sometimes represent the histone-DNA complex (called a "nucleosome") in
that way, as in the following drawing of three nucleosomes and various
associated molecules:
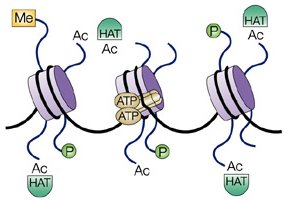 |
| From http://www.nature.com. |
It may therefore come as a surprise to you, as it once did for me, to see
an attempt at a more detailed representation of a nucleosome, as conjured
by the remarkable imaging technologies of recent years:
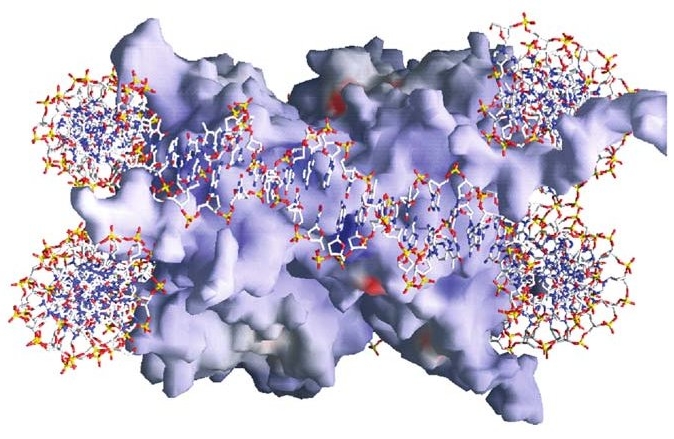 Surface image of a nucleosome with enwrapped
DNA. From Luger 2006.
Surface image of a nucleosome with enwrapped
DNA. From Luger 2006. |
For clarity's sake, the DNA here is shown merely as a red, white, yellow, and blue stick model. Arriving from elsewhere — very likely from another nearby nucleosome — the DNA meets the "spool" at upper right, wraps around the back of it, circles diagonally downward across the front face, and then around the back again, exiting at lower left toward a third (also not shown) nucleosome. The red areas of both the histone complex and the DNA are acidic and negatively charged, while the blue areas are basic and positively charged.
The spool is compounded of eight histone proteins (two each of four different types), and the DNA adheres to it by means of some 240 direct and indirect points of contact (Luger 2006). Various electrical forces play a huge role here. There is, for example, an electrostatic attraction between the largely positive surface of the histone spool and the negatively charged outer parts of the spiraling double helix. (You can see the predominance of red in the circumferential region of the DNA cross-section at lower left.)
Yet this description still invites huge misconceptions. After all, we have yet to encounter any structures in the cell nucleus that are not preeminently functioning, dynamic forms, and the nucleosome proves no different. The static image shown above, valuable as it is, becomes a lie as soon as we take it at face value.
I have struggled for a long time with the question, How can one approach such an image in the most truthful way? I suspect there is no very good answer at the current stage of our understanding. But we can at least reckon with the lesson of twentieth-century physics: there's nothing but trouble when we imagine our theoretical entities and constructs at the submicroscopic level as if they were "made of" anything like the matter of our everyday experience. At the atomic and molecular level our descriptions have more to do with centers of force and the intricate play of forces than with anything like the physical stuff of our common experience. And if this is true, then any graphic depiction of a nucleosome must be an attempt to hint at the momentary "shape" and equilibrium of innumerable intersecting forces — not the form of something like an infinitesimal lump of clay. The interactions of these forces with our sophisticated instrumentation — and not the images we unavoidably form based on our routine perceptions of the macroscopic world — are all we know of the molecular realm.
To populate that realm with mere passive "stuff" — including the kind of stuff we usually think of as constituting machines — is to render all reasonable thinking about it impossible. The literature is in fact full of references to machines — "nucleosome remodeling machines", "chromatin-modifying machines", "DNA translocation machines", "general transcription machinery", "molecular machines", and much more. The usage occurs for no discernible reason and never with even a token attempt to connect the phenomena under investigation with the machines of our experience — as opposed, say, to a connection with rocks or clouds or emotions or governments or anything else. The thoughtlessness of it all is not very becoming to the scientist who otherwise is engaged in an extraordinarily rigorous analysis requiring great discipline and caution in the use of descriptive terms.
Somewhere between 75 and 90 percent of our DNA![]() is wrapped around nucleosomes
is wrapped around nucleosomes![]() (Segal et
al. 2006), and this DNA is, as a rule, less accessible than "naked DNA"
(Segal et
al. 2006), and this DNA is, as a rule, less accessible than "naked DNA"![]() to the
various proteins that bind
to the
various proteins that bind![]() to it and help direct its performance.
This raises a question: how does the enwrapped
DNA become accessible to all the relevant binding factors, and, when it
comes time to transcribe
to it and help direct its performance.
This raises a question: how does the enwrapped
DNA become accessible to all the relevant binding factors, and, when it
comes time to transcribe![]() a gene into
mRNA
a gene into
mRNA![]() , how does
the transcription complex move along the histone
, how does
the transcription complex move along the histone![]() -bound double
helix
-bound double
helix![]() and "unzip"
its two strands? If DNA holds a crucial store of information for the
functioning of the organism, it seems odd that millions of nucleosomes
— necessary as they might be for the compaction of DNA within the
cell nucleus — should put the greater part of that information more
or less out of reach.
and "unzip"
its two strands? If DNA holds a crucial store of information for the
functioning of the organism, it seems odd that millions of nucleosomes
— necessary as they might be for the compaction of DNA within the
cell nucleus — should put the greater part of that information more
or less out of reach.
But the nucleosome, it turns out, is far from being a mere passive obstacle to DNA access. Not only has it proven to be extraordinarily plastic, mobile, and changeable, but it also plays a central role in determining what genes are "allowed" to do. You might think of it as a kind of gatekeeper, standing between the definitive textual clarity of the genomic "dictionary", on the one hand, and the entire collection of more diffuse cellular processes, on the other hand — processes seeking to compose their interwoven stories by drawing on the expressive resources of the inherited dictionary.
If there's any one place where gene regulation comes to its clearest
focus, bringing together nearly all the epigenetic processes we've been
considering, it's at the nucleosome.
Nucleosome
Positioning. To begin with, the mere presence of a nucleosome![]() at a
particular place can either repress or encourage a gene's expression
at a
particular place can either repress or encourage a gene's expression![]() . Before a
gene can be transcribed
. Before a
gene can be transcribed![]() , various
transcription factors
, various
transcription factors![]() ,
co-activators
,
co-activators![]() , and other
proteins, as well as the molecular complexes involved in the actual
transcription, must be able to bind
, and other
proteins, as well as the molecular complexes involved in the actual
transcription, must be able to bind![]() to the appropriate regulatory DNA
to the appropriate regulatory DNA![]() sequences
sequences![]() . If a
nucleosome occupies any one of those sites — that is, if the
regulatory sequence is wrapped around the nucleosome — the site can
become unavailable to the factors required for transcription.
Alternatively, the absence of a nucleosome at regulatory sites can
increase a gene's accessibility.
. If a
nucleosome occupies any one of those sites — that is, if the
regulatory sequence is wrapped around the nucleosome — the site can
become unavailable to the factors required for transcription.
Alternatively, the absence of a nucleosome at regulatory sites can
increase a gene's accessibility.
The positioning of nucleosomes matters at a highly refined level: a shift
in position as little as two or three base pairs![]() can make the
difference between an expressed or silenced
can make the
difference between an expressed or silenced![]() gene
(Martinez-Campa 2004). It also can alter the combination of other
regulatory factors necessary for gene expression. For example, the
presence or absence of a nucleosome near a gene's promoter
gene
(Martinez-Campa 2004). It also can alter the combination of other
regulatory factors necessary for gene expression. For example, the
presence or absence of a nucleosome near a gene's promoter![]() (its primary
regulatory site, generally located immediately upstream
(its primary
regulatory site, generally located immediately upstream![]() from the
gene itself) may determine whether a particular combination of
activators
from the
gene itself) may determine whether a particular combination of
activators![]() is required
for transcription
is required
for transcription![]() initiation
(Morse 2007).
initiation
(Morse 2007).
In the human genome, many actively expressed genes show a distinctive
arrangement of nucleosomes around the transcription start site
where gene transcription begins — an arrangement that includes a
"nucleosome free region" and is therefore conducive to the binding![]() of the transcribing enzyme, RNA
polymerase
of the transcribing enzyme, RNA
polymerase![]() (Schones
2008). Beyond that, different classes of gene tend to have characteristic
nucleosome positioning arrangements (Ioshikhes et al. 2006). These
positions can be roughly predicted from the underlying DNA sequence, but
the exceptions are greater than the rule, and they have everything to do
with the patterns of gene expression.
(Schones
2008). Beyond that, different classes of gene tend to have characteristic
nucleosome positioning arrangements (Ioshikhes et al. 2006). These
positions can be roughly predicted from the underlying DNA sequence, but
the exceptions are greater than the rule, and they have everything to do
with the patterns of gene expression.
A generally held view today is that DNA sequences provide a pool of
possibilities for nucleosome positions, while various regulatory factors
select among these possibilities according to the requirements for gene
expression (Vinayachandran et al. 2009; Pennings et al. 2005). Very
recent research shows a pattern of nucleosome positioning that (at least
in a broad, statistical manner) strongly diverges between genes that are
more or less continuously expressed and those whose expression varies
greatly depending on environmental conditions. The key regulatory DNA
sequences for continuously expressed genes tend to be unfriendly for
nucleosomes and therefore nucleosome-free, whereas the regulatory
sequences for the variably expressed genes are commonly occupied by
nucleosomes (Tirosh and Barkai 2008; Choi and Kim 2009). In the latter
case, the regulation of gene expression![]() is achieved
by means of all the factors (see below) that position, relocate, or
otherwise affect nucleosomes.
is achieved
by means of all the factors (see below) that position, relocate, or
otherwise affect nucleosomes.
How does the DNA sequence influence nucleosome positioning? In an
unconstrained state, DNA would not bend in anything like the degree
required in order for it to wrap around a nucleosome spool. Remember the
negatively charged outer region of the double helix![]() : a bend
would bring some of the negative charges above and below the bend into
closer proximity. The repulsion between the two charged regions will
resist the bending, as will certain interactions between the nucleotide
bases
: a bend
would bring some of the negative charges above and below the bend into
closer proximity. The repulsion between the two charged regions will
resist the bending, as will certain interactions between the nucleotide
bases![]() in the
interior of the double helix. So a powerful shifting and rebalancing of forces is
required in order to bend the DNA around the nucleosome spool. And it
happens that certain base sequences
in the
interior of the double helix. So a powerful shifting and rebalancing of forces is
required in order to bend the DNA around the nucleosome spool. And it
happens that certain base sequences![]() of the DNA
lend themselves to this readjustment more than others. In fact, the
affinity of nucleosome spools for different DNA sequences can vary by
several orders of magnitude (Pennings et al. 2005).
of the DNA
lend themselves to this readjustment more than others. In fact, the
affinity of nucleosome spools for different DNA sequences can vary by
several orders of magnitude (Pennings et al. 2005).
The many implications of nucleosome positioning remain to be worked out,
and they involve subtleties I have ignored. For example, another
epigenetic process — DNA methylation![]() (discussed
in Part 1) — also has a "voice" in positioning
nucleosomes. The general pattern of nucleosome positions varies from one
cell type to another, and distinctive patterns correlate with particular
diseases. And nucleosomes not only occlude DNA binding sites
(discussed
in Part 1) — also has a "voice" in positioning
nucleosomes. The general pattern of nucleosome positions varies from one
cell type to another, and distinctive patterns correlate with particular
diseases. And nucleosomes not only occlude DNA binding sites![]() , with a
repressive effect upon transcription; they can also bring together two
regulatory sites that need to interact in order for transcription to
occur. They do this by wrapping up the length of DNA separating the two
sites (Zhao et al. 2001).
, with a
repressive effect upon transcription; they can also bring together two
regulatory sites that need to interact in order for transcription to
occur. They do this by wrapping up the length of DNA separating the two
sites (Zhao et al. 2001).
The nucleosome's intimate association with DNA takes the form of an
elaborate complementation and reconciliation of form and force, well
calculated in some cases for longer-term stability and in others, as we
will see, for rapid readjustment in the interests of gene expression![]() or
silencing
or
silencing![]() .
.
Nucleosome Sliding. Nucleosomes![]() do not just
"sit there". One aspect of their dynamism is this: the nucleosome spool
can slide back and forth along the double helix
do not just
"sit there". One aspect of their dynamism is this: the nucleosome spool
can slide back and forth along the double helix![]() . Or,
putting it the other way around: the enwrapped double helix can slide
around the spool in one direction or another. This movement, especially
at promoter
. Or,
putting it the other way around: the enwrapped double helix can slide
around the spool in one direction or another. This movement, especially
at promoter![]() sites, can
either expose or conceal critical regulatory sequences
sites, can
either expose or conceal critical regulatory sequences![]() on the DNA
on the DNA![]() , thereby encouraging or inhibiting a
gene's transcription
, thereby encouraging or inhibiting a
gene's transcription![]() . And,
depending on the overall pattern of nucleosome positions, a section of
chromatin
. And,
depending on the overall pattern of nucleosome positions, a section of
chromatin![]() may either
wind up more compactly or else unwind and become more accessible. Without
nucleosome mobility, the dynamism of chromosomes
may either
wind up more compactly or else unwind and become more accessible. Without
nucleosome mobility, the dynamism of chromosomes![]() , so
important for gene regulation (see Part
2), would be impossible.
, so
important for gene regulation (see Part
2), would be impossible.
How does the sliding occur? Not much is known, beyond the fact that
numerous protein molecules — chromatin remodeling complexes![]() , about which
we will hear more shortly — can assist in the repositioning.
Presumably such complexes bind to the histone
, about which
we will hear more shortly — can assist in the repositioning.
Presumably such complexes bind to the histone![]() spool and
also to the DNA, applying force to pull the DNA around. Depending on the
remodeling complex, the process may sometimes involve first loosening the
DNA from the spool, and can also proceed via partial dismantling of the
spool. In some cases it may be that nucleosomes are "stably remodeled"
into a more fluent state, so that they can slide at elevated rates for a
time even without the further action of remodeling complexes. This would
also mean that their DNA is more readily accessible to transcription
factors
spool and
also to the DNA, applying force to pull the DNA around. Depending on the
remodeling complex, the process may sometimes involve first loosening the
DNA from the spool, and can also proceed via partial dismantling of the
spool. In some cases it may be that nucleosomes are "stably remodeled"
into a more fluent state, so that they can slide at elevated rates for a
time even without the further action of remodeling complexes. This would
also mean that their DNA is more readily accessible to transcription
factors![]() (Cosgrove et
al. 2004).
(Cosgrove et
al. 2004).
In Part 2 we saw the chromosome as a mobile, living entity, moving meaningfully within the living cell. Now we can amplify this picture. The spools that do so much to give the chromosome its structure are themselves mobile and living, subject to continual rearrangement. Their movements are profoundly meaningful, helping to shape the cell's effective responses to its environment. The nucleosome, writes biologist Karen Arndt in Nature, participates in "an intricate balance" and is a "dynamic structure that regulates almost all aspects of DNA metabolism" (Arndt 2007).
But our full appreciation of this flexible dynamism will require a rather
more detailed look.
Chromatin Remodeling Complexes. We
heard a good deal about transcription factors![]() in Part
1 of this series. They bind
in Part
1 of this series. They bind![]() directly to DNA
directly to DNA![]() , requiring a particular sequence
, requiring a particular sequence![]() of
nucleotide bases
of
nucleotide bases![]() in order to
do so. This enables them to play a direct role in the regulation of
individual genes, inhibiting or enhancing their expression
in order to
do so. This enables them to play a direct role in the regulation of
individual genes, inhibiting or enhancing their expression![]() .
.
There is another, very different, large, and diverse class of proteins
that are generally not sequence-specific, but that nevertheless have a
profound effect upon gene expression. They commonly bind to nucleosomes![]() , bringing
cellular stores of energy to bear upon them, and thereby participate in
positive or negative regulation of gene expression. In general, these
chromatin remodeling complexes
, bringing
cellular stores of energy to bear upon them, and thereby participate in
positive or negative regulation of gene expression. In general, these
chromatin remodeling complexes![]() either slide
the nucleosome's histone
either slide
the nucleosome's histone![]() spool along
the DNA, or else alter the spool's composition. The compositional
alterations can include replacement of some of the histones in the spool
with variant
spool along
the DNA, or else alter the spool's composition. The compositional
alterations can include replacement of some of the histones in the spool
with variant![]() histones
(see below); loosening of DNA from the spool (which makes the DNA more
accessible to other regulatory factors); or, in conjunction with
chaperone
histones
(see below); loosening of DNA from the spool (which makes the DNA more
accessible to other regulatory factors); or, in conjunction with
chaperone![]() proteins,
the ejection of one or more spool histones (Schnitzler 2008; Jiang and
Pugh 2009). While remodeling complexes do not target exact DNA
sequences
proteins,
the ejection of one or more spool histones (Schnitzler 2008; Jiang and
Pugh 2009). While remodeling complexes do not target exact DNA
sequences![]() , they can be
recruited by transcription factors
, they can be
recruited by transcription factors![]() that
are sequence-specific.
that
are sequence-specific.
A chromatin remodeling complex is indeed a complex: subtraction or
addition of proteins or other chemical groups can dramatically alter the
overall effect of the complex. One subunit of the complex, for example,
might enable it to target specific histone modifications![]() of the sort
we spoke about in Part 1 — or to make such modifications itself.
Another subunit might make it possible for a transcription factor to
recruit the complex in order to help make a gene accessible for
transcription — or to silence the gene.
of the sort
we spoke about in Part 1 — or to make such modifications itself.
Another subunit might make it possible for a transcription factor to
recruit the complex in order to help make a gene accessible for
transcription — or to silence the gene.
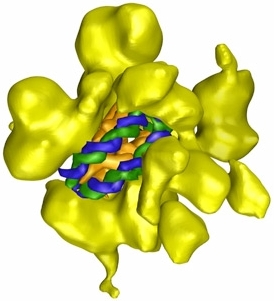 Illustration from http://www.scripps.edu/news/sr/sr2007/cb07asturias.html |
I show here one researcher's representation of a particular chromatin
remodeling complex known as "RSC". This protein is from yeast, but has
close analogs in mammals and humans. Possessing seventeen protein
subunits, it is shown as yellow in the illustration. The nucleosome is
bound in the central cavity of the RSC, with the spool shown in gold, and
the blue-and-green DNA helix![]() wrapped
twice around the spool.
wrapped
twice around the spool.
The interactions between the RSC and DNA of the nucleosome are thought to
destabilize the histone spool and loosen the DNA from it, allowing
nucleosome sliding![]() or spool
modification. This, of course, requires movement — probably the
pulling of free DNA onto the spool in the form of a large loop, which then
is passed around the spool. But it is hard to imagine the actual play of
force and movement involved, based on the kind of images and models we are
given. We can, however, at least remind ourselves of this necessary play
by, first, keeping in mind the dynamic equilibrium of thousands of
interpenetrating centers of force, alluded to at the beginning of this
article, and second, by noting that RSC has been "observed" in at least
two distinct conformations (shown below) and somehow must get from one to
the other.
or spool
modification. This, of course, requires movement — probably the
pulling of free DNA onto the spool in the form of a large loop, which then
is passed around the spool. But it is hard to imagine the actual play of
force and movement involved, based on the kind of images and models we are
given. We can, however, at least remind ourselves of this necessary play
by, first, keeping in mind the dynamic equilibrium of thousands of
interpenetrating centers of force, alluded to at the beginning of this
article, and second, by noting that RSC has been "observed" in at least
two distinct conformations (shown below) and somehow must get from one to
the other.
You will notice that the difference between the two conformations is not
simply a matter of a "lever" being switched between alternative positions
(although that is how it is often described). As with any equilibrium of
forces, a change in one place alters the entire constellation. No part of
the first structure remains exactly the same in the second. We're looking
at a set of plastic potentials, and somehow, out of this well-directed
plasticity, the necessary engagement of forces and the productive movement
does occur. According to the team of scientists who produced the two
images below, "RSC appears to be able to translocate several hundreds of
base pairs![]() at an
average velocity of 12 base pairs per second" (Leschziner et al. 2007).
at an
average velocity of 12 base pairs per second" (Leschziner et al. 2007).
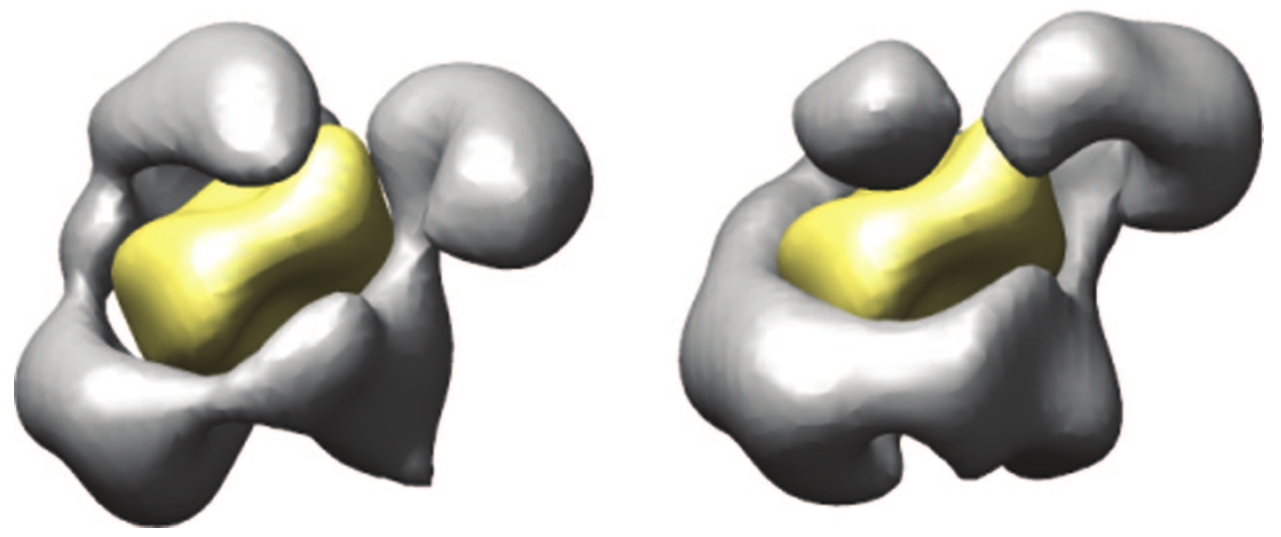 Illustration from Leschziner et al. 2007. |
Molecular biologists Cassandra Hogan and Patrick Varga-Weisz of the
Babraham Institute in Cambridge, UK, provide a hint of the interplay
between a constellation such as RSC and the larger context. They describe
how the addition or subtraction of protein subunits can alter the
functioning of these remodeling complexes. Speaking of one particular
subunit, they tell us that its addition to certain larger complexes (1)
increases the ability of the complexes to assemble regularly spaced
nucleosome![]() arrays; (2)
enhances nucleosome sliding
arrays; (2)
enhances nucleosome sliding![]() efficiency;
(3) changes the direction in which the complex moves a nucleosome; (4)
changes which histone tails
efficiency;
(3) changes the direction in which the complex moves a nucleosome; (4)
changes which histone tails![]() are
necessary for the activity of the complex; and (5) targets the complex to
particular sites within chromatin (Hogan and Varga-Weisz 2007) — all
with direct effects upon gene regulation.
are
necessary for the activity of the complex; and (5) targets the complex to
particular sites within chromatin (Hogan and Varga-Weisz 2007) — all
with direct effects upon gene regulation.
There is scarcely space to allude to many relevant aspects of chromatin remodeling complexes. They can make nucleosome arrays more evenly spaced, contributing to compaction of the chromosome, or they can disorder these arrays (Schnitzler 2008). They can recruit or interact with countless other proteins, and they can in turn be recruited by other proteins. It's been estimated that there are hundreds or even thousands of distinct remodeling complexes, each one having its own intricately sculpted, plastic form, each one interacting with selected patterns in the DNA sequence according to its own "interests", and each one potentially targeting a different set of nucleosomes for unique repositioning or alteration. Different remodeling complexes can move the same nucleosome to different positions, while a single complex might perform diverse actions upon nucleosomes at different DNA sites, thereby "allowing for intricate, gene-specific chromatin transitions" (Schnitzler 2008; also see Rippe et al. 2007).
In sum, the marriage of nucleosome to DNA appears to be every bit as
complex as the genome![]() itself, and
chromatin remodeling proteins play a big part in making it all work. And
yet we have hardly begun to survey the range of subtle gestural potentials
of the nucleosome.
itself, and
chromatin remodeling proteins play a big part in making it all work. And
yet we have hardly begun to survey the range of subtle gestural potentials
of the nucleosome.
Histone Modifications. If we could
actually see the play of form and force at the level of the nucleosome,
perhaps we would be most impressed with the long, relatively unstructured,
and highly mobile histone tails![]() . Comprising
some 25-30% of the mass of the nucleosome spool, these active, sinuous
tails can form contacts with the encircling DNA, helping to regulate its
binding to the spool and its accessibility to transcription factors
. Comprising
some 25-30% of the mass of the nucleosome spool, these active, sinuous
tails can form contacts with the encircling DNA, helping to regulate its
binding to the spool and its accessibility to transcription factors![]() . But with a
shift of their "attention" they can also link up with nearby protein
constituents of the chromatin and, by doing so, contribute to the
compaction of the chromatin fiber (Zheng and Hayes 2003). In addition,
they can be enablers or disablers of the work of remodeling complexes.
. But with a
shift of their "attention" they can also link up with nearby protein
constituents of the chromatin and, by doing so, contribute to the
compaction of the chromatin fiber (Zheng and Hayes 2003). In addition,
they can be enablers or disablers of the work of remodeling complexes.
The main body of research on the tails, however, has focused on the
"histone modifications"![]() I spoke of
in Part 1 — the attachment or detachment of
(mostly) small chemical groups that, in countless combinations, can
decorate the histone tails. Among the modifiers are the methyl
I spoke of
in Part 1 — the attachment or detachment of
(mostly) small chemical groups that, in countless combinations, can
decorate the histone tails. Among the modifiers are the methyl![]() , acetyl
, acetyl![]() , and
phosphate
, and
phosphate![]() groups, as
well as the small protein, ubiquitin
groups, as
well as the small protein, ubiquitin![]() . By their
means the cell marks
. By their
means the cell marks![]() its
nucleosomes — perhaps less forcefully than is accomplished by
chromatin remodeling proteins
its
nucleosomes — perhaps less forcefully than is accomplished by
chromatin remodeling proteins![]() , but with
equally wide-ranging effects upon gene expression
, but with
equally wide-ranging effects upon gene expression![]() .
.
Karolin Luger of the Howard Hughes Medical Institute in Maryland could
already write in 2006 that some 150 distinct histone modifications were
known, and a steady stream of new ones have been identified since then.
Whereas such modifications were at first thought to occur mainly on the
histone tails, many have more recently been found on the core histones
themselves, where they become targets attracting particular proteins,
including the remodeling complexes![]() .
.
These histone modifications are virtually all reversible, and the proteins
responsible for their attachment and removal "are likely present at the
same time", leading to "constant dynamic change" (Luger 2006).
Acetylation![]() ,
methylation
,
methylation![]() , and
phosphorylation
, and
phosphorylation![]() can appear
or disappear within minutes of the arrival of an appropriate stimulus at
the cell surface (Kouzarides 2007). One modification can influence the
occurrence of other modifications on the same histone or even on other
nucleosomes (Altaf 2009). Some modifications, or their combinations, are
strongly associated with gene expression
can appear
or disappear within minutes of the arrival of an appropriate stimulus at
the cell surface (Kouzarides 2007). One modification can influence the
occurrence of other modifications on the same histone or even on other
nucleosomes (Altaf 2009). Some modifications, or their combinations, are
strongly associated with gene expression![]() while others
are associated with gene repression
while others
are associated with gene repression![]() . Some
localize at gene promoters
. Some
localize at gene promoters![]() while others
mark the body of the gene. Some occur in tightly packed chromatin and
others in unwound chromatin. Some predominate in particular regions of
the nucleus.
while others
mark the body of the gene. Some occur in tightly packed chromatin and
others in unwound chromatin. Some predominate in particular regions of
the nucleus.
Whereas the chromatin remodeling complexes account for much of the
movement and restructuring of nucleosomes, histone modifications are often
thought of more as signals — signals that, among other things, give
the remodeling complexes their cues for action. There is surely truth in
this, but it also needs remembering that these modifying chemical groups,
small as they may be compared to the massive protein complexes,
nevertheless can subtly shift structural balances, with immediate and
dramatic consequences. Histone modifications, by altering the shape and
charge of nucleosomes, can change the accessibility of DNA![]() , the ease of nucleosome sliding
, the ease of nucleosome sliding![]() , and the
packing of chromatin
, and the
packing of chromatin![]() .
.
But, of course, the more indirect, signaling functions of the ever-shifting histone modifications remain vitally important. They enable the nucleosome to become a flexible target for numerous remodeling and regulatory factors capable of recognizing the modifications. It is often difficult to disentangle the supposed direct effects of a modification from those others brought about through its role in convoking a larger assembly of convergent factors (Choi and Howe 2009). And, moreover, an undue compulsion to disentangle can sometimes appear slightly perverse, as if there were a wish to reduce the contextual cell to a more comprehensible collection of isolatable causes and effects — causes and effects whose description is always misleading because such isolation never occurs in the living cell.
But, however you look at it, the changing pattern of histone![]() marks
marks![]() modulates — and is in turn
modulated by — almost every genetic and epigenetic process in the
cell. In this way the changing needs of the cell come to a focus at the
nucleosome and are reconciled with the structure and expressive potentials
of DNA.
modulates — and is in turn
modulated by — almost every genetic and epigenetic process in the
cell. In this way the changing needs of the cell come to a focus at the
nucleosome and are reconciled with the structure and expressive potentials
of DNA.
A fairly simple example may help. In 2007 Andrea Smallwood from the
school of medicine at UCLA, along with her colleagues, published a paper
on the relation between one particular histone mark![]() and DNA methylation
and DNA methylation![]() . DNA
methylation, you may recall from Part
1, involves the application of methyl groups
. DNA
methylation, you may recall from Part
1, involves the application of methyl groups![]() , not to
histones or their tails, but rather directly to DNA — and
specifically to the cytosine bases
, not to
histones or their tails, but rather directly to DNA — and
specifically to the cytosine bases![]() of DNA.
Such methylation — at least when it occurs in the promoter
of DNA.
Such methylation — at least when it occurs in the promoter![]() region of
genes — is commonly associated with gene silencing.
region of
genes — is commonly associated with gene silencing.
One question that interested Smallwood and the others is how DNA methylation is actually guided. I will briefly summarize the proposal their research led them to in one particular sort of case. I draw here particularly on a review of this research by Carmen Brenner and François Fuks (2007), who present the illustration below, acknowledging that it is "simplistic". (Feel free to skim over the explanatory details in the bullet list.)
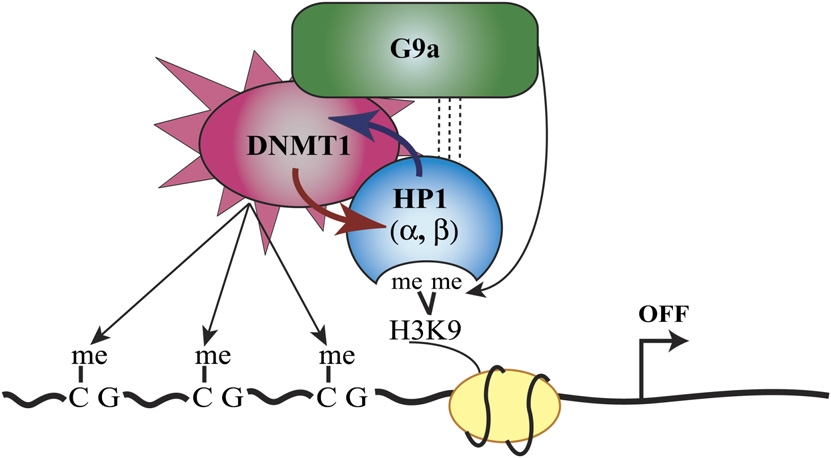 Illustration from Brenner and Fuks (2007). |
In summary, Brenner and Fuks comment that there is no single direction of
causation here between DNA and histones, but rather a "complex interplay
between mutually influencing marks![]() ". Everything varies "according to
context", resulting in "a conversation full of subtle inflections, with
multiple partners and mediations" (Brenner and Fuks 2007).
". Everything varies "according to
context", resulting in "a conversation full of subtle inflections, with
multiple partners and mediations" (Brenner and Fuks 2007).
Histone Variants and Histone Chaperones. There is one last aspect
of the nucleosome's dynamism I would like to mention. Nucleosome
sliding![]() along the
DNA molecule, the loosening or tightening of DNA-histone
along the
DNA molecule, the loosening or tightening of DNA-histone![]() contacts,
and the modification
contacts,
and the modification![]() of the
nucleosome by methyl
of the
nucleosome by methyl![]() , acetyl
, acetyl![]() , and other
chemical groups are only part of the story. It turns out that the core
spool, consisting of eight histone proteins (two each of four different
types) is itself subject to continual histone exchange, with variant
, and other
chemical groups are only part of the story. It turns out that the core
spool, consisting of eight histone proteins (two each of four different
types) is itself subject to continual histone exchange, with variant![]() histones
sometimes taking the place of canonical ones. Each variant histone can
have its own effects upon the nucleosome's role in gene regulation
histones
sometimes taking the place of canonical ones. Each variant histone can
have its own effects upon the nucleosome's role in gene regulation![]() .
.
Researchers are intrigued, for example, by a particular variant known as
H2A.Z, which is deposited by a chromatin remodeling protein![]() and tends to
show up in nucleosomes around many gene promoters
and tends to
show up in nucleosomes around many gene promoters![]() . H2A.Z
destabilizes nucleosomes, making them more susceptible to sliding —
an effect that is accentuated in the presence of a particular histone
tail
. H2A.Z
destabilizes nucleosomes, making them more susceptible to sliding —
an effect that is accentuated in the presence of a particular histone
tail![]() modification
(Schones 2008). The regulatory situation around the promoters thus
becomes more fluid and therefore can more readily be shifted between
active and inactive states in response to contextual needs. And this
fluidity is further increased when, along with H2A.Z, the variant H3.3
histone replaces the canonical H3.
modification
(Schones 2008). The regulatory situation around the promoters thus
becomes more fluid and therefore can more readily be shifted between
active and inactive states in response to contextual needs. And this
fluidity is further increased when, along with H2A.Z, the variant H3.3
histone replaces the canonical H3.
Or, anyway, that is one part of the story. But, going in something like
the opposite direction, it appears that H2A.Z can also help to form
compact chromatin![]() , rendering
DNA less accessible. It all depends on the larger circumstances (Altaf et
al. 2009).
, rendering
DNA less accessible. It all depends on the larger circumstances (Altaf et
al. 2009).
While the investigation of histone variants![]() is still at
an early stage, there is no shortage of interesting observations, such as
the fact that the length of DNA wrapped around a nucleosome containing
H2A.Z is considerably shorter than the length around a "normal" nucleosome
(Tolstorukov et al. 2009) — thereby changing the availability of
binding
is still at
an early stage, there is no shortage of interesting observations, such as
the fact that the length of DNA wrapped around a nucleosome containing
H2A.Z is considerably shorter than the length around a "normal" nucleosome
(Tolstorukov et al. 2009) — thereby changing the availability of
binding![]() sequences
sequences![]() . Or the
fact that the loss of another variant, H2A.X, "compromises genomic
integrity and increases cancer incidence" under certain conditions
(Hadnagy et al. 2008). Or the fact that histones are rapidly exchanged
even in most regions of densely compacted chromatin, contributing to the
general plasticity of chromatin (Luger 2006). And, in the kind of
circular causation we've become so familiar with in this review, not only
do certain remodeling complexes
. Or the
fact that the loss of another variant, H2A.X, "compromises genomic
integrity and increases cancer incidence" under certain conditions
(Hadnagy et al. 2008). Or the fact that histones are rapidly exchanged
even in most regions of densely compacted chromatin, contributing to the
general plasticity of chromatin (Luger 2006). And, in the kind of
circular causation we've become so familiar with in this review, not only
do certain remodeling complexes![]() mediate the
substitution of variant histones for canonical ones, but the variant
histones in turn seem to help regulate the remodeling complexes —
for example, by limiting their ability to condense chromatin (Hogan and
Varga-Weisz 2007).
mediate the
substitution of variant histones for canonical ones, but the variant
histones in turn seem to help regulate the remodeling complexes —
for example, by limiting their ability to condense chromatin (Hogan and
Varga-Weisz 2007).
Remodeling complexes can do more than exchange histone constituents; they
can also evict one or more of the histones, leaving various sorts of
"incomplete" spool. This contributes even more radically to a free, open,
and accessible state of DNA, while also facilitating nucleosome sliding![]() . Remodeling
of this sort can proceed all the way to complete nucleosome disassembly.
. Remodeling
of this sort can proceed all the way to complete nucleosome disassembly.
The reconstruction of nucleosomes is facilitated by other molecules beside
chromatin remodelers. A large and diverse group of protein complexes
called "histone chaperones"![]() is
continually in attendance upon histones, seeing to their assembly, their
deposition upon DNA, their proper interactions with proteins, and their
disassembly. Chaperones not only participate in the displacement of
particular histones from nucleosomes; they can remove the entire spool,
leaving naked DNA
is
continually in attendance upon histones, seeing to their assembly, their
deposition upon DNA, their proper interactions with proteins, and their
disassembly. Chaperones not only participate in the displacement of
particular histones from nucleosomes; they can remove the entire spool,
leaving naked DNA![]() . Prior
destabilization of a nucleosome by a variant histone can make the removal
easier.
. Prior
destabilization of a nucleosome by a variant histone can make the removal
easier.
As you might expect by this point, other cellular and epigenetic processes
bear on nucleosome stability. For example, histone modifications![]() such as
acetylation
such as
acetylation![]() facilitate
nucleosome eviction (Schones 2008). Micro-RNAs
facilitate
nucleosome eviction (Schones 2008). Micro-RNAs![]() (discussed
in Part 1) conduce to the formation of compact chromatin
and therefore are implicated in the stabilization of nucleosomes.
Positive supercoiling
(discussed
in Part 1) conduce to the formation of compact chromatin
and therefore are implicated in the stabilization of nucleosomes.
Positive supercoiling![]() ahead of the
transcribing enzyme
ahead of the
transcribing enzyme![]() facilitates
removal of certain histones from the nucleosome spool (Zlatanova 2009).
And, finally, DNA binding sites
facilitates
removal of certain histones from the nucleosome spool (Zlatanova 2009).
And, finally, DNA binding sites![]() for
transcription factors
for
transcription factors![]() also play a
role:
also play a
role:
Specific transcription factor-binding sites have been found to correlate with nucleosome eviction in vivo, suggesting that certain transcription factor complexes may gain access to DNA by excluding nucleosomes. The latter is consistent with in vitro
studies illustrating that nucleosomes can be destabilized or excluded by cooperative binding of transcription factors". (Leimgruber et al. 2009)
A Histone Code? Because of the remarkable array of histone
modifications![]() and the
ongoing torrent of discoveries about distinct effects resulting from
different combinations of them, some researchers have argued for a
precise, combinatorial histone code
and the
ongoing torrent of discoveries about distinct effects resulting from
different combinations of them, some researchers have argued for a
precise, combinatorial histone code![]() ,
recognizable by chromatin remodeling complexes
,
recognizable by chromatin remodeling complexes![]() and all
sorts of chromatin and DNA binding
and all
sorts of chromatin and DNA binding![]() factors. The idea is that each unique
combination of modifications could be neatly "read off" by the appropriate
factors as dictating a specific action.
factors. The idea is that each unique
combination of modifications could be neatly "read off" by the appropriate
factors as dictating a specific action.
But the idea of a code implies a syntactic fixity that simply has never
been found. For example, different proteins, by responding to the same
histone modification, can make it a signal for either transcriptional
activation![]() or acute
gene silencing
or acute
gene silencing![]() (Rando and
Chang 2009). One would think the epigenetic fate of that supposedly
quintessential and most definitive of codes — the all-explaining
"Master Plan" we were supposed to find in the DNA
(Rando and
Chang 2009). One would think the epigenetic fate of that supposedly
quintessential and most definitive of codes — the all-explaining
"Master Plan" we were supposed to find in the DNA![]() sequence
sequence![]() —
would have cured us of the yearning for such a conveniently well-defined
syntax governing the living creature.
—
would have cured us of the yearning for such a conveniently well-defined
syntax governing the living creature.
Where is the need for such a code, anyway? No one can doubt that these histone modifications speak; they mean something. They both carry messages from, and issue messages to, the cellular environment, so that the nucleosome's mediating role in relation to gene expression can be carried out. It's just that the meaning of these words must be read, like all meaningful text, within a larger context that never remains precisely what it was before. As one molecular biologist has put it, "The more we look in to [histone] modifications, the more it will become clear that context is everything" (Kouzarides 2007).
What we see here is what we see everywhere in the organism: no single set of conditions ever speaks categorically, independent of a yet wider range of conditions. That's why one forever encounters statements like this in the literature of gene regulation: "While some studies suggest that DNA methylation patterns guide histone modifications (including histone acetylation and methylation) during gene silencing, other studies argue that DNA methylation takes its cues primarily from histone modification states" (Vaissière et al. 2008). So which is it: A causes B or B causes A? In an organic context the question makes no sense, except as a useful stimulus to focus one's vision on detail in an approximate and provisional manner.
It only needs adding that the right sort of contextual understanding remains elusive. Researchers continually report new connections between the "bewildering array" of histone modifications (Rando and Chang, 2009) and all sorts of other epigenetic goings-on. The details, as you've now had a chance to glimpse, have become rather overwhelming in their luxuriant diversity. But it's not at all clear that anyone is getting much closer to putting it all together in a satisfying manner.
Before trying to place the nucleosome as meaningfully as possible within its larger context, I would like to offer a brief prediction.
Within a year or two some highly placed researcher, secure enough in his or her position of authority to take the risk, will publish a dramatic statement to the following effect:
What are we doing? Every month we gather more data on the genome and epigenome in an ever-rising flood. We learn more and more details about more and more minute processes, and the dizzying pace of discovery provokes use of the word "exciting" in one technical paper after another. But has no one noticed that we seem to be getting farther and farther away from an understanding of cell and organism?We used to have a clear framework for saying what made what happen. DNA gave us a blueprint and a First Cause to which everything else could be traced in a hierarchical fashion. At the top of the hierarchy was a single set of crystal-precise molecules, and somewhere below was everything else we see in the living organism.
That blueprint, however, has disappeared. What is there to take its place? The satisfyingly clear lines of cause and effect are, with every exciting new discovery, dissolving further into a chaos of causal arrows pointing in all possible directions. Where are the higher-level ordering principles? Yes, we clearly are gaining countless useful facts, but is there anything causal, anything explanatory, holding these facts together in the way that the organism itself so obviously holds together?
This, of course, leaves open the decisive question: What sort of approach would manifest the unity of the organism itself? That's an inquiry for the later articles in this series. But it may not be premature to point out that the question seems to contain a pointer to its own answer. That is, it expresses a recognition of the unity of the organism. We must have gained that recognition somehow. Maybe the task before us is simply to pursue such recognition more fully in its own natural terms. Maybe the reason our search for machine-like explanations loses the organism is that the organism is not machine-like.
At the conclusion of this excursion through the dynamic regulatory
landscape of the nucleosome![]() , we can
hardly help agreeing with the judgment of the researcher who, in a classic
1997 paper, published the definitive "crystal structure of the nucleosome
core particle" (and who is responsible for the image at the beginning of
this article). She has more recently written:
, we can
hardly help agreeing with the judgment of the researcher who, in a classic
1997 paper, published the definitive "crystal structure of the nucleosome
core particle" (and who is responsible for the image at the beginning of
this article). She has more recently written:
It has become clear that the many cellular activities that impinge upon chromatin structure operate via multiple and complex mechanisms. Mounting evidence demonstrates that ATP-dependent chromatin remodelling factors
, histone-chaperones
, histone modifying
enzymes, and nucleosome-binding proteins affect different levels of chromatin
organization in a highly orchestrated and concerted manner. It is highly unlikely that, for example, every promoter
is made accessible via a unified order of events; rather, each and every incidence of regulated DNA
accessibility will have to be studied independently to identify the important players and order of events that are necessary for the regulation of DNA accessibility. (Luger 2006)
Another prominent molecular biologist, acknowledging that "we might have been much too rigid in thinking about how nucleosomes function", reminds us that "virtually every aspect" of the nucleosome is subject to modification and dynamic change. There was evidence of this, she notes, "from the earliest days", but it was "generally neglected" in favor of "a 'fixture' in our minds, an artifactual entity" created as a result of particular experimental and environmental conditions (Zlatanova et al. 2009). I would only add that deeply entrenched habits of thought can also constrain the researcher's vision. The Hungarian physiologist Albert Szent-Györgyi is reported to have said, "Discovery consists in seeing what everybody has seen and thinking what nobody has thought."
So how can we begin to think about the nucleosome and all the
processes it is caught up in? And how can we do so without losing
ourselves in all the details — that is, without feeling oppressed by
what easily becomes a meaningless juxtaposition of "one damned thing after
another"? (I suspect you've cottoned on to this challenge by now!)
Probably we're a long way from having any sort of coherent, contextual
picture — something it would be very healthy for biologists to
acknowledge now and then. The only thing I know to do in such a situation
is to step back a little and seek a broader perspective.
The
Nucleosome as Mediator. In the living organism we always find
ourselves confronting relatively fixed structure and organization, on the
one hand, and plastic energies on the other. The cell nucleus![]() presents us
with structure most vividly in the given sequence of DNA bases
presents us
with structure most vividly in the given sequence of DNA bases![]() .
Plasticity, on the other hand, comes into play through chromosome looping,
remodeling, and the entire range of epigenetic processes we've been
looking at, by which the organism adjusts to its environment. Samuel
Taylor Coleridge spoke of an irreducible polarity between "confining form"
and "free life", and it is indeed impossible to have life without
structure, identity, and fixed form, just as it is is also impossible to
have life without movement, flexibility, and change. The movement needs
the fixed structure to "play off of". Muscles would be of no use if there
were not the hard skeleton to pull against.
.
Plasticity, on the other hand, comes into play through chromosome looping,
remodeling, and the entire range of epigenetic processes we've been
looking at, by which the organism adjusts to its environment. Samuel
Taylor Coleridge spoke of an irreducible polarity between "confining form"
and "free life", and it is indeed impossible to have life without
structure, identity, and fixed form, just as it is is also impossible to
have life without movement, flexibility, and change. The movement needs
the fixed structure to "play off of". Muscles would be of no use if there
were not the hard skeleton to pull against.
Within this context, I find it natural to think of the nucleosome![]() as a kind of
mediator between the relative fixity of DNA
as a kind of
mediator between the relative fixity of DNA![]() and the ceaselessly varied metabolic
flows of the cell as a whole. On the one hand, it is in the most intimate
possible contact with the stable structure of DNA, literally enwrapped
— and, one might think, entrapped — within a coil of the
double helix
and the ceaselessly varied metabolic
flows of the cell as a whole. On the one hand, it is in the most intimate
possible contact with the stable structure of DNA, literally enwrapped
— and, one might think, entrapped — within a coil of the
double helix![]() . But it
might be truer to say that the nucleosome embraces DNA, marrying
itself to and complementing the local bends, twists, and electrical
tensions of the double helix and thereby maintaining an intricate and
finely adjustable balance of forces. Certain of the filamentary histone
tails
. But it
might be truer to say that the nucleosome embraces DNA, marrying
itself to and complementing the local bends, twists, and electrical
tensions of the double helix and thereby maintaining an intricate and
finely adjustable balance of forces. Certain of the filamentary histone
tails![]() (depending
on their various modifications) actually wrap themselves around the double
helix, conforming to its spiraling grooves and helping to hold it firmly
in place.
(depending
on their various modifications) actually wrap themselves around the double
helix, conforming to its spiraling grooves and helping to hold it firmly
in place.
If that were all, the cell could not live, for the DNA would be a rigid,
unmanageable, and dead structure. But while intimately adapting itself to
the structure of DNA, the nucleosome also stands as a kind of "attractor"
for a seemingly endless range of modifying and restructuring agents
streaming in from the wider cellular environment — chromatin
remodeling complexes![]() ,
chaperones
,
chaperones![]() , histone
modifying
, histone
modifying![]() enzymes,
transcription factors
enzymes,
transcription factors![]() , and much
more. The nucleosome provides a kind of integrating center where these
often highly diverse influences — fleeting or more enduring,
insistent or more reticent — can be weighed together and allowed to
speak in a unified, harmonious voice. It is one of those places where the
larger context comes to especially vivid and confluent expression.
, and much
more. The nucleosome provides a kind of integrating center where these
often highly diverse influences — fleeting or more enduring,
insistent or more reticent — can be weighed together and allowed to
speak in a unified, harmonious voice. It is one of those places where the
larger context comes to especially vivid and confluent expression.
This middle position, where the nucleosome must reconcile the fixed
structure and expressive potentials of DNA with the vast array of actors
upon which gene expression![]() depends,
demands great dexterity. And the nucleosome is in fact capable of
remarkable movement and transformation. It can slide one way or another
so as to expose or put out of reach crucial DNA regulatory sequences
depends,
demands great dexterity. And the nucleosome is in fact capable of
remarkable movement and transformation. It can slide one way or another
so as to expose or put out of reach crucial DNA regulatory sequences![]() ; it
participates centrally in the higher-order structuring — the
condensation and decondensation — of DNA; it allows its activity to
be continually modulated by the rich repertoire of histone modifications
; it
participates centrally in the higher-order structuring — the
condensation and decondensation — of DNA; it allows its activity to
be continually modulated by the rich repertoire of histone modifications![]() ; it can be
repeatedly disassembled and then reassembled in varying degrees, as is
required in a carefully orchestrated manner during gene transcription
; it can be
repeatedly disassembled and then reassembled in varying degrees, as is
required in a carefully orchestrated manner during gene transcription![]() ; it can
incorporate or suffer incorporation of variant
; it can
incorporate or suffer incorporation of variant![]() histones
(the distinction between actor and acted-upon is forever blurred in the
living cell), with pronounced effect upon its functioning; the filamentary
tail
histones
(the distinction between actor and acted-upon is forever blurred in the
living cell), with pronounced effect upon its functioning; the filamentary
tail![]() that in one
situation helps to bind DNA to the histone spool may, with proper
modification, serve to loosen the DNA, making the nucleosome more
malleable and the DNA more accessible to transcription factors
that in one
situation helps to bind DNA to the histone spool may, with proper
modification, serve to loosen the DNA, making the nucleosome more
malleable and the DNA more accessible to transcription factors![]() ....
....
It has become almost a cliché among molecular biologists to speak of the nucleosome as carrying a contradictory burden with regard to DNA:
The histones within the nucleosome have evolved to accomplish two conflicting yet vital tasks: first, the approximately 2 meters of . . . DNA have to be packaged within the confines of the nucleus, preventing knots and tangles and protecting the genome from physical damage. Second, the information that is encoded within the DNA needs to be accessed at appropriate times, and this is to a large part regulated by local changes in nucleosome and chromatin structure by complex mechanisms that are only now emerging. (Luger 2006)
One certainly understands what she means. At the same time, it is well to note that the nucleosome shows no sign of being conflicted or torn in opposite directions. It deftly mediates between the transient requirements of a Protean environment on one side, and the intrinsic character or "givenness" of tightly packed DNA on the other. If life manifests itself as a dynamic equilibrium between limitation and plasticity (Holdrege 1996), surely the nucleosome gives us an especially vivid picture of this equilibrium. It is Proteus himself — though with his feet rooted firmly in the "solid ground" of DNA.
The nucleosome's meaningful and effective shifts of form seem to present
no less a challenge to our understanding than the developing form of the
organism as a whole — if only we were in a position to see and
interpret the modulations of form and force that constitute its gesturing!
The Nucleosome's Dance. I have just now suggested that
the role of a mediator requires flexible powers of adjustment, adaptation,
and movement. But, more than that, effective mediation demands grace and
rhythm if one is to hold an interactive balance between contrasting
demands. These qualities, it seems to me, can at least be glimpsed from a
distance in the nucleosome.
Certain processes offer particularly suggestive images of the flexibility
and grace required of the nucleosome. One of the most thoroughly studied
genes is PHO5 in yeast. Researchers have recently found that when
this particular gene is active, roughly two of the three nucleosomes on
its promoter![]() are removed,
on average (Boeger et al. 2008). I say "roughly" because the evidence
suggests a thoroughly dynamic state of affairs. It appears that the outer
two nucleosomes are continually dissassembled and reassembled, while the
central nucleosome remains intact — intact, but not immobile.
are removed,
on average (Boeger et al. 2008). I say "roughly" because the evidence
suggests a thoroughly dynamic state of affairs. It appears that the outer
two nucleosomes are continually dissassembled and reassembled, while the
central nucleosome remains intact — intact, but not immobile.
What happens, according to the researchers, is that the intact nucleosome,
assisted by the RSC chromatin remodeling complex![]() ,
repetitively slides over the promoter, first in one direction and then the
other. In the process, it successively interacts with each of the other
two nucleosomes, dislodging their histone
,
repetitively slides over the promoter, first in one direction and then the
other. In the process, it successively interacts with each of the other
two nucleosomes, dislodging their histone![]() spools from
the DNA or, perhaps, simply sliding them out of the way. A dislodged
nucleosome may quickly be reassembled when the central actor moves in the
opposite direction. By this means different regions of the promoter are
alternately exposed to the succession of protein complexes necessary for
repeated transcription
spools from
the DNA or, perhaps, simply sliding them out of the way. A dislodged
nucleosome may quickly be reassembled when the central actor moves in the
opposite direction. By this means different regions of the promoter are
alternately exposed to the succession of protein complexes necessary for
repeated transcription![]() of the
PHO5 gene.
of the
PHO5 gene.
Then there is what some have called "histone modification pulsing" in
human embryonic stem cells. These cells must remain "pluripotent" —
capable of differentiating![]() into various
sorts of specialized cell. The genes relating to specialized development
in stem cells must be kept silent; but they must also be in a state of
readiness for expression as soon as differentiation begins.
into various
sorts of specialized cell. The genes relating to specialized development
in stem cells must be kept silent; but they must also be in a state of
readiness for expression as soon as differentiation begins.
This carefully balanced state, it has been proposed, is facilitated by the nucleosomes associated with the DNA loci controlling differentiation. These nucleosomes seem to undergo a rapid and continual "pulsing" of seemingly contradictory histone modifications. Some of the modifications normally favor gene expression, while others favor silencing — but neither set of modifications gains the upper hand. There is a continual alternation between them, keeping the genes, so to speak, in a state of "suspended readiness". Then, when the decision to specialize is finally taken, the repressive modifications are discontinued and the genes begin to be expressed (Gan et al. 2007).
More generally, a rather different sort of equilibrium can hold in balance
the appropriate possibilities of a gene's expression. Cizhong Jiang and
Franklin Pugh (2009) suggest that an optimum mixture of favorable and
unfavorable nucleosome positioning sequences![]() can
establish a carefully weighed balance between "a state that can be
disrupted to allow transcription
can
establish a carefully weighed balance between "a state that can be
disrupted to allow transcription![]() and
replication
and
replication![]() and a stable
state that prevents inappropriate access to DNA".
and a stable
state that prevents inappropriate access to DNA".
An excellent example of this is offered by a particular remodeling complex called "Isw2". Well-studied in yeast, it has been found to move nucleosome spools "outward" from the ends of some genes onto the adjacent regulatory sequences, thereby repressing gene transcription (Whitehouse and Tsukiyama 2006; Whitehouse et al. 2007). Because the original positions have DNA sequences that are favorable for nucleosomes, whereas the positions imposed by Isw2 tend to be very unfavorable, such nucleosomes can, according to Jiang and Pugh, be thought of as "spring-loaded": as soon as Isw2 is removed, the nucleosomes return to their original, more favorable positions, allowing rapid expression of the affected genes.
There is, then (as Whitehouse and Tsukiyama put it), a tension between the "antagonistic forces of Isw2 and the DNA sequence" — a tension through which gene regulation is achieved. The ability to move with these contrasting forces and to hold the balance between them seems very much in character for the mediating nucleosome.
And, finally, in yet another sort of balancing act, the nucleosome with
its DNA is subject to what many investigators refer to as a "breathing" of
DNA on the histone![]() spool.
There is evidence that the ends of the nucleosome-bound DNA — where
the DNA enters and leaves the spool — can momentarily relax and
"peel away" from the spool, then return to it (Luger 2006; Zlatanova et
al. 2009). This rhythmic alternation, whose cycle is measured in
milliseconds, offers what are presumably well-calibrated opportunities for
regulatory factors to bind
spool.
There is evidence that the ends of the nucleosome-bound DNA — where
the DNA enters and leaves the spool — can momentarily relax and
"peel away" from the spool, then return to it (Luger 2006; Zlatanova et
al. 2009). This rhythmic alternation, whose cycle is measured in
milliseconds, offers what are presumably well-calibrated opportunities for
regulatory factors to bind![]() to otherwise inaccessible stretches of
DNA. It would be no accident, then, that nucleosomes tend to assume
positions such that DNA regulatory sequences — the sequences that
regulatory factors bind to — reside near the entry and exit sites of
the nucleosomes (Jiang and Pugh 2009).
to otherwise inaccessible stretches of
DNA. It would be no accident, then, that nucleosomes tend to assume
positions such that DNA regulatory sequences — the sequences that
regulatory factors bind to — reside near the entry and exit sites of
the nucleosomes (Jiang and Pugh 2009).
And beyond all this, it is hard to forget those long, sinuous histone
tails![]() , —
tails that might almost suggest the graceful tentacles of an octopus, now
embracing the nucleosomal DNA, now reaching out toward other histones in
order to restructure chromatin, and all the while adroitly re-shaping
their own gestures in response to modifying signals from the wider
environment.
, —
tails that might almost suggest the graceful tentacles of an octopus, now
embracing the nucleosomal DNA, now reaching out toward other histones in
order to restructure chromatin, and all the while adroitly re-shaping
their own gestures in response to modifying signals from the wider
environment.
It's possible to look from a slightly different angle at the relation
between the stable structure of DNA and the ever-shifting, kaleidoscopic
environment. It happens that the "breathing" on the nucleosome![]() of the
otherwise tightly bound double helix
of the
otherwise tightly bound double helix![]() is not the
only way that DNA can be loosened from the spool and made available to
transcription factors
is not the
only way that DNA can be loosened from the spool and made available to
transcription factors![]() and other
external regulatory agents. Certain chromatin remodeling complexes
and other
external regulatory agents. Certain chromatin remodeling complexes![]() transiently
expose DNA regulatory sequences by creating small DNA loops on the
nucleosome surface (Jiang and Pugh 2009).
transiently
expose DNA regulatory sequences by creating small DNA loops on the
nucleosome surface (Jiang and Pugh 2009).
This may remind us that spiraling and looping are fundamental gestures of
the chromosome, from the spiraling of the individual strands of the double
helix around each other, to the spiraling of DNA around the nucleosome
spool, to the spiraling of histone tails around that same DNA, to the
supercoiling![]() of longer
stretches of chromatin
of longer
stretches of chromatin![]() , to the
large looping movements we recognized in Part
2 as elements of long-distance gene regulation. And to these we can
now add the formation of small DNA loops on the nucleosome surface.
, to the
large looping movements we recognized in Part
2 as elements of long-distance gene regulation. And to these we can
now add the formation of small DNA loops on the nucleosome surface.
Circling, looping, and spiraling movements are, you might say, archetypal images of the constancy within change that characterizes all life. And between the smallest and largest of these intranuclear movements, we see the nucleosome, wrapping itself in the double helix and figuring decisively in many of the other movements, sliding one way or another, coiling or uncoiling chromatin, "sensing" the surroundings with its undulating tails, continually dissolving and reconstituting its own substance through the exchange of histones, accommodating numerous modifications and variants that re-shape its balance of forces — and all this, perhaps, while stretches of DNA are rhythmically "breathing" by lifting off its surface and settling back. And out of this fluid sculptural performance, orchestrated as it is by the cell as a whole, each of our 25,000 genes finds its appropriate expression.
(You will find the latest versions of the currently available parts of this series at the website, "From Mechanism to a Science of Qualities".)
Please Note: With a view toward the needs of the readership, I have preferred to cite review articles, where they are available and, in general, have made little effort to reflect in my citations the priority claims of the various investigators of any particular phenomenon. Public (online) accessibility of papers and ease of access to the relevant information are primary criteria for my selection — qualified, of course, by the limits of my own familiarity with the literature.
Altaf, Mohammed, Andréanne Auger, Marcela Covic et al. (2009). "Connection between Histone H2A Variants and Chromatin Remodeling Complexes", Biochemistry and Cell Biology vol. 87, pp. 35-50. doi:10.1139/O08-140
Arndt, Karen M. (2007). "Genome under Surveillance", Nature vol. 450 (Dec. 13), pp. 959-60.
Azuara, Véronique, Pascale Perry, Stephan Sauer et al. (2006). "Chromatin Signatures of Pluripotent Cell Lines", Nature Cell Biology vol. 8 no. 5 (May), pp. 532-8. doi:10.1038/ncb1403
Barski, Artem, Suresh Cuddapah, Kairong Cui et al. (2007) "High-resolution Profiling of Histone Methylations in the Human Genome", Cell vol. 129, no. 4 (May 18), pp. 823-37. doi:10.1016/j.cell.2007.05.009
Bártová, Eva, Jana Krejci, Andrea Harnicarová et al. (2008). "Histone Modifications and Nuclear Architecture: A Review", Journal of Histochemistry and Cytochemistry vol. 56, no. 8, pp. 711-21. doi:10.1369/jhc.2008.951251
Bernstein, Bradley E., Tarjei S. Mikkelsen, Xiaohui Xie et al. (2006). "A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells", Cell vol. 125 (April 21), pp. 315-26. doi:10.1016/j.cell.2006.02.041
Boeger, Hinrich, Joachim Griesenbeck, and Roger D. Kornberg (2008). "Nucleosome Retention and the Stochastic Nature of Promoter Chromatin Remodeling for Transcription", Cell vol. 133, no. 4 (May 16), pp. 716-26. doi:10.1016/j.cell.2008.02.051
Brenner, Carmen and François Fuks (2007). "A Methylation Rendezvous: Reader Meets Writer", Developmental Cell vol. 12 (June), pp. 843-4. doi:10.1016/j.devcel.2007.05.011
Choi, Jennifer K. and LeAnn J. Howe (2009). "Histone Acetylation: Truth of Consequences?" Biochemistry and Cell Biology vol. 87, pp. 139-50. doi:10.1139/O08-112
Choi, J. K. and Y.-J. Kim (2009). "Intrinsic Variability of Gene Expression Encoded in Nucleosome Positioning Sequences", Nature Genetics vol. 41 (Mar. 1), pp. 498-503. doi:10.1038/ng.319
Cosgrove, Michael S., Jef D. Boeke, and Cynthia Wolberger (2004). "Regulated Nucleosome Mobility and the Histone Code", Nature Structural and Molecular Biology, vol. 11, no. 11 (Nov.), pp. 1037-43.
Crepaldi, Luca and Antonella Riccio (2009). "Chromatin Learns to Behave", Epigenetics vol. 4, no. 1 (Jan. 1), pp. 23-6.
Fischle, Wolfgang, Boo Shan Tseng, Holger L. Dormann et al. (2005). "Regulation of HP1-chromatin Binding by Histone H3 Methylation and Phosphorylation", Nature vol. 438, no. 22 (Dec. 29), pp. 1116-22. doi:10.1038/nature04219
Gan, Qiong, Tadashi Yoshida, Oliver G. McDonald et al. (2007). "Concise Review: Epigenetic Mechanisms Contribute to Pluripotency and Cell Lineage Determination of Embryonic Stem Cells", Stem Cells vol. 25, no. 1, pp. 2-9. doi:10.1634/stemcells.2006-0383
Hadnagy, Annamaria, Raymond Beaulieu, and Danuta Balicki (2008). "Histone Tail Modifications and Noncanonical Functions of Histones: Perspectives in Cancer Epigenetics", Molecular Cancer Therapeutics vol. 7, no. 4 (April), pp. 740-8. doi:10.1158/1535-7163.MCT-07-2284
Hogan, Cassandra and Patrick Varga-Weisz (2007). "The Regulation of ATP-dependent Nucleosome Remodelling Factors", Mutation Research vol. 618, pp. 41-51. doi:10.1016/j.mrfmmm.2006.07.010
Holdrege, Craig (1996). Genetics and the Manipulation of Life: The Forgotten Factor of Context. Hudson NY: Lindisfarne.
Ioshikhes, Ilya P., Istvan Albert, Sara J. Zanton, et al. (2006). "Nucleosome Positions Predicted through Comparative Genomics", Nature Genetics vol. 38, no. 10, pp. 1210-5. doi:10.1038/ng1878
Jiang, Cizhong and B. Franklin Pugh (2009). "Nucleosome Positioning and Gene Regulation: Advances through Genomics", Nature Reviews Genetics (advance epublication). doi:10.1038/nrg2522
Kouzarides, Tony (2007). "Chromatin Modifications and Their Function", Cell vol. 128 (Feb. 23), pp. 693-705. doi:10.1016/j.cell.2007.02.005
Leimgruber, Elisa, Queralt Sequin-Estévez, Isabelle Dunand-Sauthier, et al. (2009). "Nucleosome Eviction from MHC Class II Promoters Controls Positioning of the Transcription Start Site", Nucleic Acids Research (Mar 5; epublication ahead of print). doi:10.1093/nar/gkp116
Luger, Karolin (2006). "Dynamic Nucleosomes", Chromosome Research vol. 14, pp. 5-16. doi:10.1007/s10577-005-1026-1
Martinez-Campa, Carlos, Panagiotis Politis, Jean-Luc Moreau, et al. (2004). "Precise Nucleosome Positioning and the TATA Box Dictate Requirements for the Histone H4 Tail and the Bromodomain Factor Bdf1", Molecular Cell vol. 15 (July 2), pp. 69-81.
Pennings, Sari, James Allan, and Colin S. Davey (2005). "DNA Methylation, Nucleosome Formation and Positioning", Briefings in Functional Genomics and Proteomics vol. 3, no. 4, pp. 351-61.
Rando, Oliver J. and Howard Y. Chang (2009). "Genome-Wide Views of Chromatin Structure", Annual Review of Biochemistry vol. 78 (Feb. 17), pp. 13.1-13.27. doi:10.1146/annurev.biochem.78.071107.134639
Rippe, Karsten, Anna Schrader, Philipp Riede, et al. (2007). "DNA Sequence- and Conformation-directed Positioning of Nucleosomes by Chromatin-Remodeling Complexes", PNAS vol. 104, no. 40 (Oct. 2), pp. 15635-40. doi:10.1073/pnas.0702430104
Schnitzler, Gavin R. (2008). "Control of Nucleosome Positions by DNA Sequence and Remodeling Machines", Cell Biochemistry and Biophysics vol. 51, nos. 2-3 (July), pp. 67-80. doi:10.1007/s12013-008-9015-6
Schones, Dustin E., Kairong Cui, Suresh Cuddapah, et al. (2008). "Dynamic Regulation of Nucleosome Positioning in the Human Genome", Cell vol. 132 (Mar. 7), pp. 887-98. doi:10.1016/j.cell.2008.02.022
Segal, Eran, Yvonne Fondufe-Mittendorf, Lingyi Chen, et al. (2006). "A Genomic Code for Nucleosome Positioning", Nature vol. 442, no. 7104 (Aug. 17), pp. 772-8.
Smallwood, Andrea, Pierre-Olivier Estève Sriharsa Pradhan, et al. (2007). "Functional Cooperation between HP1 and DNMT1 Mediates Gene Silencing", Genes and Development vol. 21, no. 10 (May 15), pp. 1169-78. doi:10.1101/gad.1536807
Tirosh, Itay and Naama Barkai (2008). "Two Strategies for Gene Regulation by Promoter Nucleosomes", Genome Research vol. 18, pp. 1084-91. doi:10.1101/gr.076059.108
Tolstorukov, Michael Y., Peter V. Kharchenko, Joseph A. Goldman et al. (2009). "Comparative Analysis of H2A.Z Nucleosome Organization in the Human and Yeast Genomes", Genome Research vol. 19, pp. 967-77. doi:10.1101/gr.084830.108
Vinayachandran, Vinesh, Rama-Haritha Pusarly, and Purnima Bhargava (2009). "Multiple Sequence-directed Possibilities Provide a Pool of Nucleosome Position Choices in Different States of Activity of a Gene", Epigenetics and Chromatin vol. 2, no. 4 (Mar. 16). doi:10.1186/1756-8935-2-4
Whitehouse, Iestyn and Toshio Tsukiyama (2006). "Antagonistic Forces That Position Nucleosomes in Vivo", Nature Structural and Molecular Biology vol. 13, no. 7 (July), pp. 633-40. doi:10.1038/nsmb1111
Whitehouse, Iestyn, Oliver J. Rando, Jeff Delrow, et al. (2007). "Chromatin Remodelling at Promoters Suppresses Antisense Transcription", Nature vol. 450 (Dec. 13), pp. 1031-6. doi:10.1038/nature06391
Zheng, Chunyang and Jeffrey J. Hayes (2003). "Structures and Interactions of the Core Histone Tail Domains", Biopolymers vol. 68, pp. 539-46. doi:10.1002/bip.10303
Zlatanova, Jordanka, Thomas C. Bishop, Jean-Marc Victor, et al. (2009). "The Nucleosome Family: Dynamic and Growing", Structure vol. 17 (Feb. 13), pp. 160-71.
NetFuture, a freely distributed electronic newsletter, is published and copyrighted by The Nature Institute. The editor is Stephen L. Talbott, author of Devices of the Soul: Battling for Our Selves in the Age of Machines (http://natureinstitute.org/txt/st). You may redistribute this newsletter for noncommercial purposes. You may also redistribute individual articles in their entirety, provided the NetFuture url and this paragraph are attached.
NetFuture is supported by freely given reader contributions, and could not survive without them. For details and special offers, see http://netfuture.org/support.html .
Current and past issues of NetFuture are available on the Web:
http://netfuture.org
To subscribe or unsubscribe, go to
http://netfuture.org/subscribe.html.If you have problems subscribing or unsubscribing, send mail to: owner-netfuture@lists.panix.com.
This issue of NetFuture: http://netfuture.org/2009/Jul0909_177.html.
Steve Talbott :: NetFuture #177 :: July 9, 2009